|
Disruption of Ca2+/calmodulin:KSR1 interaction lowers ERK activation
Thines, L; Jang, H; Li, Z et al.
Protein Sci
, DOI://10.1002/pro.4982 (2024)
 MYCN drives oncogenesis by cooperating with the histone methyltransferase G9a and the WDR5 adaptor to orchestrate global gene transcription
MYCN drives oncogenesis by cooperating with the histone methyltransferase G9a and the WDR5 adaptor to orchestrate global gene transcription
Liu, Z; Zhang, X; Xu, M et al.
PLoS Biol
, DOI://10.1371/journal.pbio.3002240 (2024)
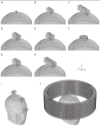 Electric Field Characteristics of Rotating Permanent Magnet Stimulation
Electric Field Characteristics of Rotating Permanent Magnet Stimulation
Robins, PL; Makaroff, SN; Dib, M et al.
Bioengineering (Basel)
, DOI://10.3390/bioengineering11030258 (2024)
 Dynamic extracellular vestibule of human SERT: Unveiling druggable potential with high-affinity allosteric inhibitors
Dynamic extracellular vestibule of human SERT: Unveiling druggable potential with high-affinity allosteric inhibitors
Salomon, K; Abramyan, AM; Plenge, P et al.
Proc Natl Acad Sci U S A
, DOI://10.1073/pnas.2304089120 (2023)
 Conserved Noncoding Elements Evolve Around the Same Genes Throughout Metazoan Evolution
Conserved Noncoding Elements Evolve Around the Same Genes Throughout Metazoan Evolution
Gonzalez, P; Hauck, QC; Baxevanis, AD; ,
Genome Biol Evol
, DOI://10.1093/gbe/evae052 (2024)
Flipped C-Terminal Ends of APOA1 Promote ABCA1-Dependent Cholesterol Efflux by Small HDLs
He, Y; Pavanello, C; Hutchins, PM et al.
Circulation
, DOI://10.1161/CIRCULATIONAHA.123.065959 (2024)
A new method for multiancestry polygenic prediction improves performance across diverse populations
Zhang, H; Zhan, J; Jin, J et al.
Nat Genet
, DOI://10.1038/s41588-023-01501-z (2023)
Comprehensive mapping of cell fates in microsatellite unstable cancer cells supports dual targeting of WRN and ATR
Zong, D; Koussa, NC; Cornwell, JA et al.
Genes Dev
, DOI://10.1101/gad.351085.123 (2023)
|
 All Services Operational
All Services Operational All Services Operational
All Services Operational