From the gopeaks documentation:
GoPeaks is a peak caller designed for CUT&TAG/CUT&RUN sequencing data. GoPeaks by default works best with narrow peaks such as H3K4me3 and transcription factors. However, broad epigenetic marks like H3K27Ac/H3K4me1 require different the step, slide, and minwidth parameters.
Allocate an interactive session and run the program.
Sample session (user input in bold) for running the peakfinder with a control and generating a summary plot with deeptools:
[user@biowulf]$ sinteractive --cpus-per-task=6 --gres=lscratch:20 --mem=30g
salloc.exe: Pending job allocation 46116226
salloc.exe: job 46116226 queued and waiting for resources
salloc.exe: job 46116226 has been allocated resources
salloc.exe: Granted job allocation 46116226
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3144 are ready for job
[user@cn3144 ~]$ module load gopeaks
[user@cn3144 ~]$ cd /lscratch/$SLURM_JOB_ID
[user@cn3144 ~]$ cp $GOPEAKS_TEST_DATA/* .
[user@cn3144 ~]$ ls -lh
[user@cn3144 ~]$ gopeaks -b GSE190793_Kasumi_cutrun.bam -c GSE190793_Kasumi_IgG.bam --mdist 1000 --prefix Kasumi_cnr
Reading chromsizes from bam header...
nTests: 431593
nzSignals: 4.5114732e+07
nzBins: 8298786
n: 7.566515e+06
p: 7.184688090883924e-07
mu: 5.962418894299423
var: 5.962414610487421
[user@cn3144 ~]$ cat Kasumi_cnr_gopeaks.json
{
"gopeaks_version": "1.0.0",
"date": "2023-05-12 12:40:59 PM",
"elapsed": "7m19.045741504s",
"prefix": "Kasumi_cnr",
"command": "gopeaks -b GSE190793_Kasumi_cutrun.bam -c GSE190793_Kasumi_IgG.bam --mdist 1000 --verbose --prefix=Kasumi_cnr",
"peak_counts": 29393
}
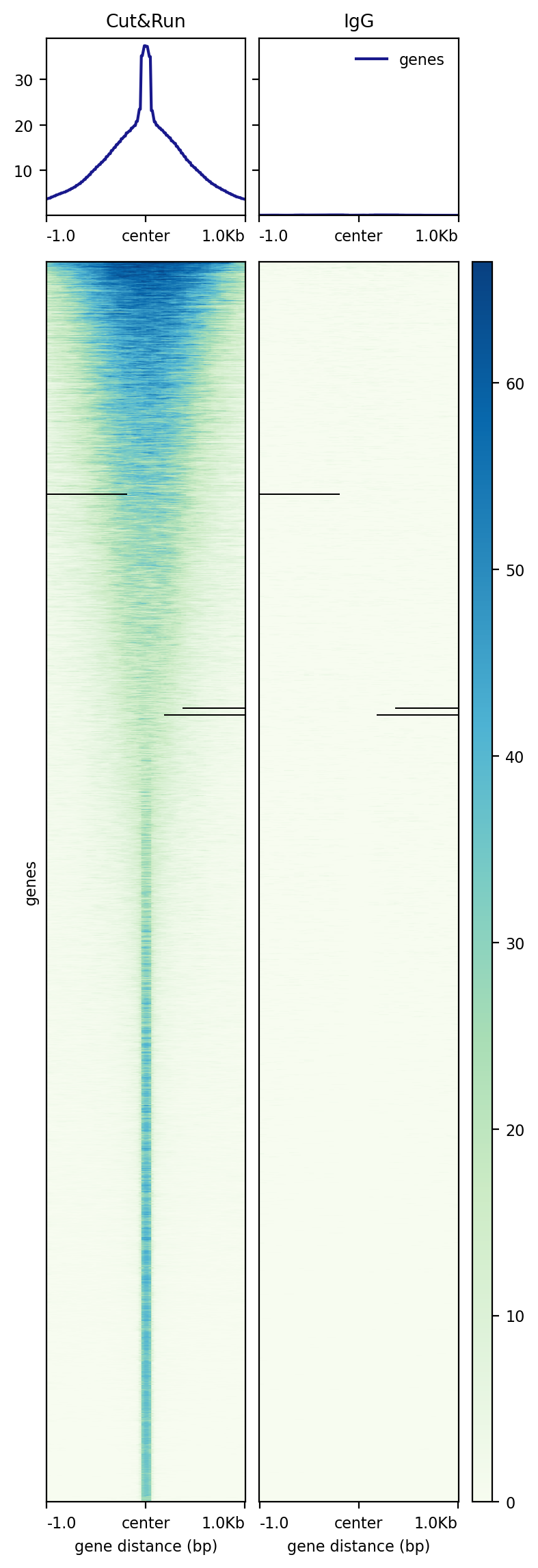
[user@cn3144 ~]$ # create a summary graph for peaks centered on the middle of the peak interval +- 1000
[user@cn3144 ~]$ # i.e. not gene annotation
[user@cn3144 ~]$ module load deeptools
[user@cn3144 ~]$ bamCoverage -p6 -b GSE190793_Kasumi_cutrun.bam -o GSE190793_Kasumi_cutrun.bw
[user@cn3144 ~]$ bamCoverage -p6 -b GSE190793_Kasumi_IgG.bam -o GSE190793_Kasumi_IgG.bw
[user@cn3144 ~]$ computeMatrix reference-point -R Kasumi_cnr_peaks.bed -a 1000 -b 1000 --referencePoint center \
-S GSE190793_Kasumi_cutrun.bw GSE190793_Kasumi_IgG.bw \
--sortRegions descend --samplesLabel 'Cut&Run' 'IgG' -p6 -o cutrun_matrix
[user@cn3144 ~]$ plotHeatmap -m cutrun_matrix -o cutrun.png --averageTypeSummaryPlot mean --colorMap GnBu
[user@cn3144 ~]$ cp Kasumi_cnr_* *.bw cutrun.png /data/$USER/my_working_directory
[user@cn3144 ~]$ exit
salloc.exe: Relinquishing job allocation 46116226
[user@biowulf ~]$

Create a batch input file (e.g. gopeaks.sh). For example:
#!/bin/bash set -e module load gopeaks/1.0.0 cp $GOPEAKS_TEST_DATA/* . gopeaks -b GSE190793_Kasumi_cutrun.bam -c GSE190793_Kasumi_IgG.bam --mdist 1000 --prefix Kasumi_cnr
Submit this job using the Slurm sbatch command.
sbatch --cpus-per-task=6 --mem=30g gopeaks.sh
Create a swarmfile (e.g. gopeaks.swarm). For example:
gopeaks -b replicate1.bam -c control.bam --mdist 1000 --prefix replicate1_gopeaks gopeaks -b replicate2.bam -c control.bam --mdist 1000 --prefix replicate2_gopeaks
Submit this job using the swarm command.
swarm -f gopeaks.swarm -g 30 -t 6 --module gopeakswhere
| -g # | Number of Gigabytes of memory required for each process (1 line in the swarm command file) |
| -t # | Number of threads/CPUs required for each process (1 line in the swarm command file). |
| --module gopeaks | Loads the gopeaks module for each subjob in the swarm |