From the Treemix site:
TreeMix is a method for inferring the patterns of population splits and mixtures in the history of a set of populations. In the underlying model, the modern-day populations in a species are related to a common ancestor via a graph of ancestral populations. We use the allele frequencies in the modern populations to infer the structure of this graph.
The input is a gzipped file with a header containing a space-delimited list of the names of populations, followed by lines containing the allele counts at each SNP. The order of the SNPs in the file is assumed to reflect the order of the SNPs in the genome. The line is space delimited between populations, and the two allele within the population are comma-delimited. For example:
pop1 pop2 pop3 pop4 5,1 1,1 4,0 0,4 3,3 0,2 2,2 0,4 1,5 0,2 2,2 1,3
$TREEMIX_TEST_DATAAllocate an interactive session and run the program. Sample session:
[user@biowulf]$ sinteractive
salloc.exe: Pending job allocation 46116226
salloc.exe: job 46116226 queued and waiting for resources
salloc.exe: job 46116226 has been allocated resources
salloc.exe: Granted job allocation 46116226
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3144 are ready for job
[user@cn3144]$ module load treemix
[user@cn3144]$ #copy example data to local directory
[user@cn3144]$ cp ${TREEMIX_TEST_DATA:-none}/treemix_test_files.tar.gz .
[user@cn3144]$ tar -xzf treemix_test_files.tar.gz
[user@cn3144]$ ls -lh
total 820K
-rw-r----- 1 user group 196 Feb 17 2012 pop_order_test
-rw-r----- 1 user group 402K Feb 17 2012 testin.gz
[user@cn3144]$ zcat testin.gz | head -n5
Han Sardinian Colombian Dai French Mozabite Karitiana Lahu BiakaPygmy She Italian San Yoruba
2,66 2,54 0,10 1,19 4,52 6,48 0,24 2,14 31,13 0,20 3,23 6,6 21,21
11,55 2,54 0,10 1,19 4,52 2,52 0,26 3,13 10,36 2,18 1,25 5,7 12,32
12,56 2,54 0,10 1,19 4,52 2,52 0,26 3,13 10,36 2,18 1,25 5,7 12,32
0,68 0,56 0,10 0,20 0,56 1,53 0,26 0,16 9,37 0,20 0,26 2,10 10,32
[user@cn3144]$ treemix -i testin.gz -o testout
TreeMix v. 1.12
$Revision: 231 $
npop:13 nsnp:29999
Estimating covariance matrix in 29999 blocks of size 1
SEED: 1455041772
Starting from:
(Lahu:0.00323769,(San:0.0494469,She:0.0102143):0.00323769);
Adding French [4/13]
[...snip...]
[user@cn3144]$ ls -lh
total 848K
-rw-r----- 1 user group 196 Feb 17 2012 pop_order_test
-rw-r----- 1 user group 402K Feb 17 2012 testin.gz
-rw-rw---- 1 user group 735 Feb 9 13:16 testout.cov.gz
-rw-rw---- 1 user group 688 Feb 9 13:16 testout.covse.gz
-rw-rw---- 1 user group 250 Feb 9 13:16 testout.edges.gz
-rw-rw---- 1 user group 117 Feb 9 13:16 testout.llik
-rw-rw---- 1 user group 722 Feb 9 13:16 testout.modelcov.gz
-rw-rw---- 1 user group 247 Feb 9 13:16 testout.treeout.gz
-rw-rw---- 1 user group 630 Feb 9 13:16 testout.vertices.gz
[user@cn3144]$ # copy file with r functions for plotting trees
[user@cn3144]$ cp /usr/local/apps/treemix/1.12/bin/plotting_funcs.R .
[user@cn3144]$ module load R
[user@cn3144]$ R
R version 3.2.3 (2015-12-10) -- "Wooden Christmas-Tree"
Copyright (C) 2015 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
[...snip...]
> source("plotting_funcs.R")
> png(file="testout.png", width=1024, height=1024, res = 1024/7)
> plot_tree("testout", o="pop_order_test")
[...snip...]
> dev.off()
> q()
[user@cn3144]$ exit
salloc.exe: Relinquishing job allocation 46116226
[user@biowulf]$
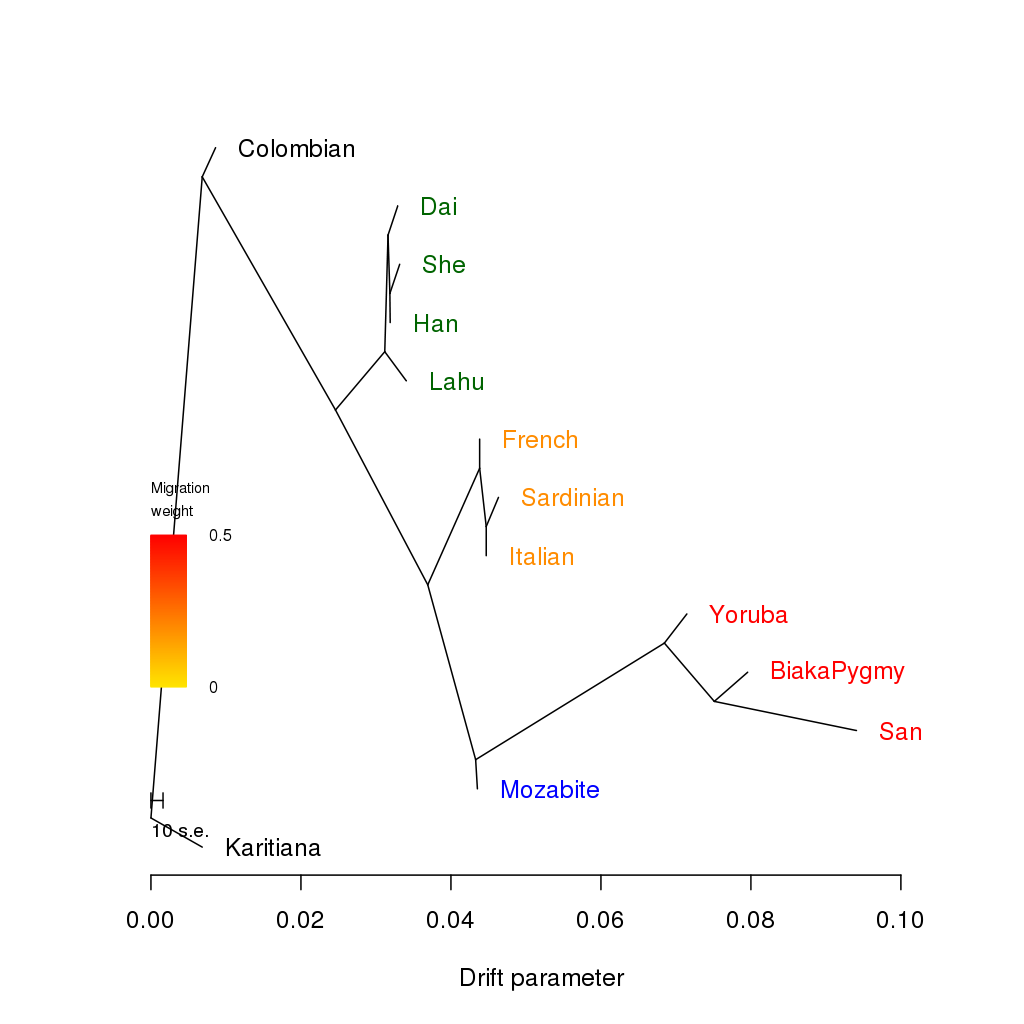
Creates the following tree

Create a batch input file (e.g. treemix.sh). For example:
#! /bin/bash module load treemix/1.12 || exit 1 treemix -i testin.gz -o testout -m 2 -global
Submit this job using the Slurm sbatch command.
sbatch --cpus-per-task=2 --mem=2g treemix.sh
Create a swarmfile (e.g. treemix.swarm). For example to run bootstrap replicates of a tree:
treemix -i testin.gz -m 2 -global -bootstrap -k 500 -o bootstrap1 treemix -i testin.gz -m 2 -global -bootstrap -k 500 -o bootstrap2 treemix -i testin.gz -m 2 -global -bootstrap -k 500 -o bootstrap3
Submit this job using the swarm command.
swarm -f treemix.swarm -g 2 -t 1 -p 2 --module treemix/1.12where
| -g # | Number of Gigabytes of memory required for each process (1 line in the swarm command file) |
| -t # | Number of threads/CPUs required for each process (1 line in the swarm command file). |
| --module treemix | Loads the treemix module for each subjob in the swarm |