CellRank is a modular framework to study cellular dynamics based on Markov state modeling of multi-view single-cell data. CellRank scales to large cell numbers, is fully compatible with the scverse ecosystem, and is easy to use. In the backend, it is powered by the pyGPCCA package.
Allocate an interactive session and run the program. Sample session:
[user@biowulf]$ sinteractive --mem=4g
[user@cn0911 ~]$module load scdrs
[+] Loading singularity 3.10.5 on cn4185
[+] Loading cellrank 2.0.0
[user@cn0911 ~]$mkdir /data/$USER/cellrank && cd /data/$USER/cellrank
[user@cn0911 ~]$python-cr
>>> import sys
>>> import cellrank as cr
>>> import scanpy as sc
>>> sc.settings.set_figure_params(frameon=False, dpi=100)
>>> cr.settings.verbosity = 2
>>> import warnings
>>> warnings.simplefilter("ignore", category=UserWarning)
>>> adata = cr.datasets.pancreas()
100%|█████████████████████████████████████████████████████████| 33.5M/33.5M [00:01<00:00, 18.7MB/s]
>>> adata
AnnData object with n_obs × n_vars = 2531 × 27998
obs: 'day', 'proliferation', 'G2M_score', 'S_score', 'phase', 'clusters_coarse', 'clusters', 'clusters_fine', 'louvain_Alpha', 'louvain_Beta', 'palantir_pseudotime'
var: 'highly_variable_genes'
uns: 'clusters_colors', 'clusters_fine_colors', 'day_colors', 'louvain_Alpha_colors', 'louvain_Beta_colors', 'neighbors', 'pca'
obsm: 'X_pca', 'X_umap'
layers: 'spliced', 'unspliced'
obsp: 'connectivities', 'distances'
>>> import scvelo as scv
>>> scv.pl.proportions(adata)
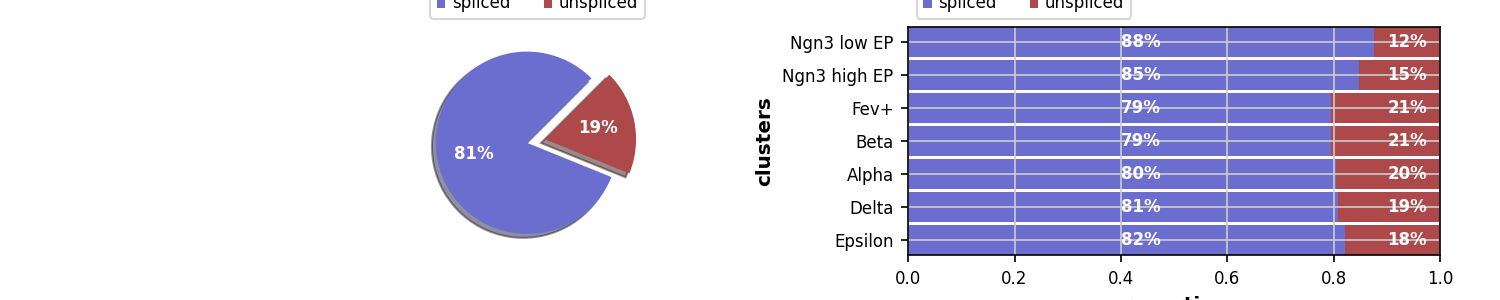
>>> scv.pp.filter_and_normalize(adata, min_shared_counts=20, n_top_genes=2000, subset_highly_variable=False)
Normalized count data: X, spliced, unspliced.
Extracted 2000 highly variable genes.
Logarithmized X.
>>> sc.tl.pca(adata)
[user@cn0911 ~]$sc.pp.neighbors(adata, n_pcs=30, n_neighbors=30, random_state=0)
[user@cn0911 ~]$scv.pp.moments(adata, n_pcs=None, n_neighbors=None)
computing moments based on connectivities
finished (0:00:01) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
[user@cn0911 ~]$sc.pl.embedding(adata, basis="umap", color=["clusters", "proliferation"])
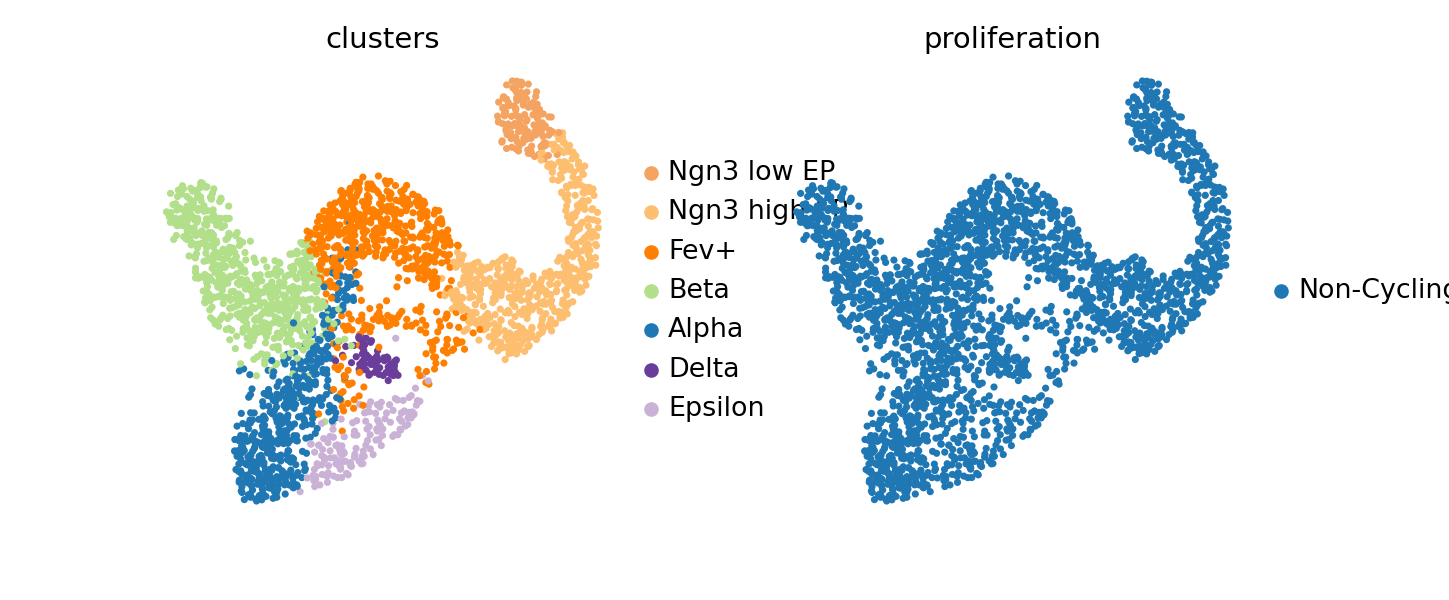 [user@cn0911 ~]$exit
End the interactive session:
[user@cn0911 ~]$exit
End the interactive session:
[user@cn0911 ~]$ exit salloc.exe: Relinquishing job allocation 46116226 [user@biowulf ~]$