Scipion is an image processing framework to obtain 3D models of macromolecular complexes using Electron Microscopy (3DEM). It integrates several software packages and presents an unified interface for both biologists and developers. Scipion allows to execute workflows combining different software tools, while taking care of formats and conversions. Additionally, all steps are tracked and can be reproduced later on.
There may be multiple versions of Scipion available. An easy way of selecting the version is to use modules. To see the modules available, type
module avail scipion
To select a module, type
module load scipion/[ver]
where [ver] is the version of choice.
Scipion is a package management framework. Much of what Scipion does is controlled via the local scipion.conf file. This is located in ~/.config/scipion:
module load scipion scipion config cat ~/.config/scipion/scipion.conf
This file is created when Scipion is first used. The SCIPION_USER_DATA parameter in the DIRS_LOCAL section of this file should be edited to direct where generated files are written, as by default it will write to your /home directory:
[DIRS_LOCAL] SCIPION_USER_DATA = /data/$USER/apps/scipion/ScipionUserData SCIPION_LOGS = %(SCIPION_USER_DATA)s/logs SCIPION_TMP = %(SCIPION_USER_DATA)s/tmp
$USER should be changed to your username.
Once an interactive session is allocated, e.g.
sinteractive --cpus-per-task=16 --mem=24g --gres=lscratch:50 --time=8:00:00
Scipion can be run like so:
module load scipion scipion tests em.workflows.test_workflow_spiderMDA
Scipion can be run as a GUI. This requires an X11 session.
scipion

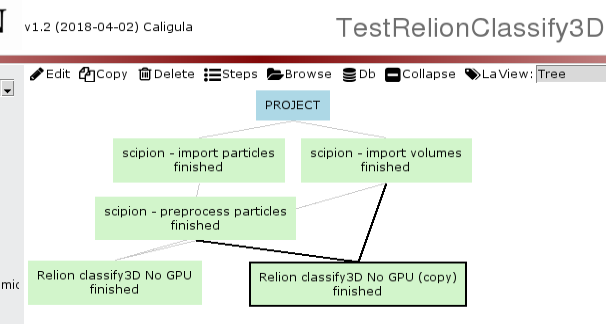
A batch job can be launched from the GUI like this:
Double click on the project step:
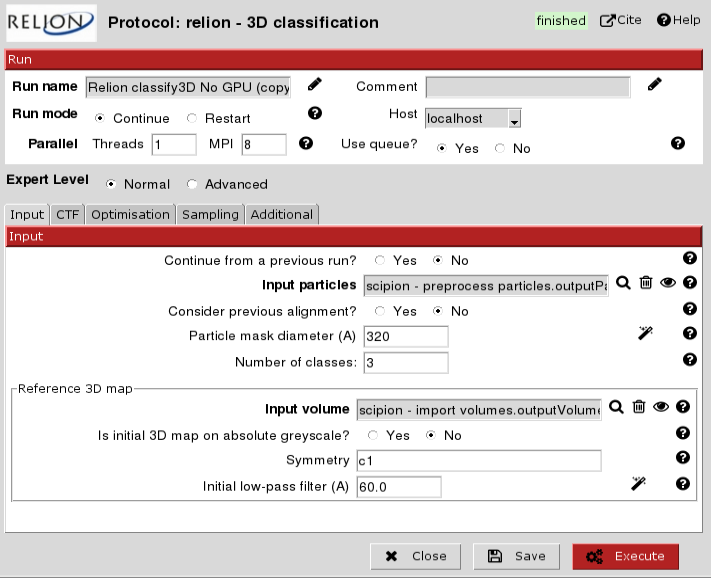
Making sure all the input parameters are correct, change 'Use queue?' from 'No' to 'Yes', then click 'Execute':
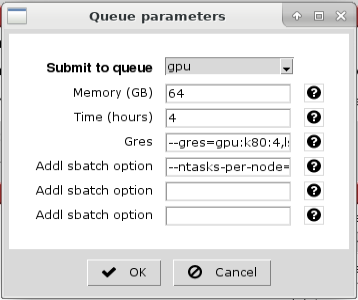
The Queue parameters window will appear. Choose the appropriate partition (queue):
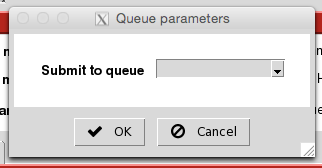
Make sure the memory, time and local scratch space are adequate, then click 'OK':
Create a batch input file (e.g. scipion.sh):
#!/bin/bash module load scipion scipion tests em.workflows.test_workflow_spiderMDA
Submit this job using the Slurm sbatch command.
sbatch --cpus-per-task=16 --mem=24g scipion.sh