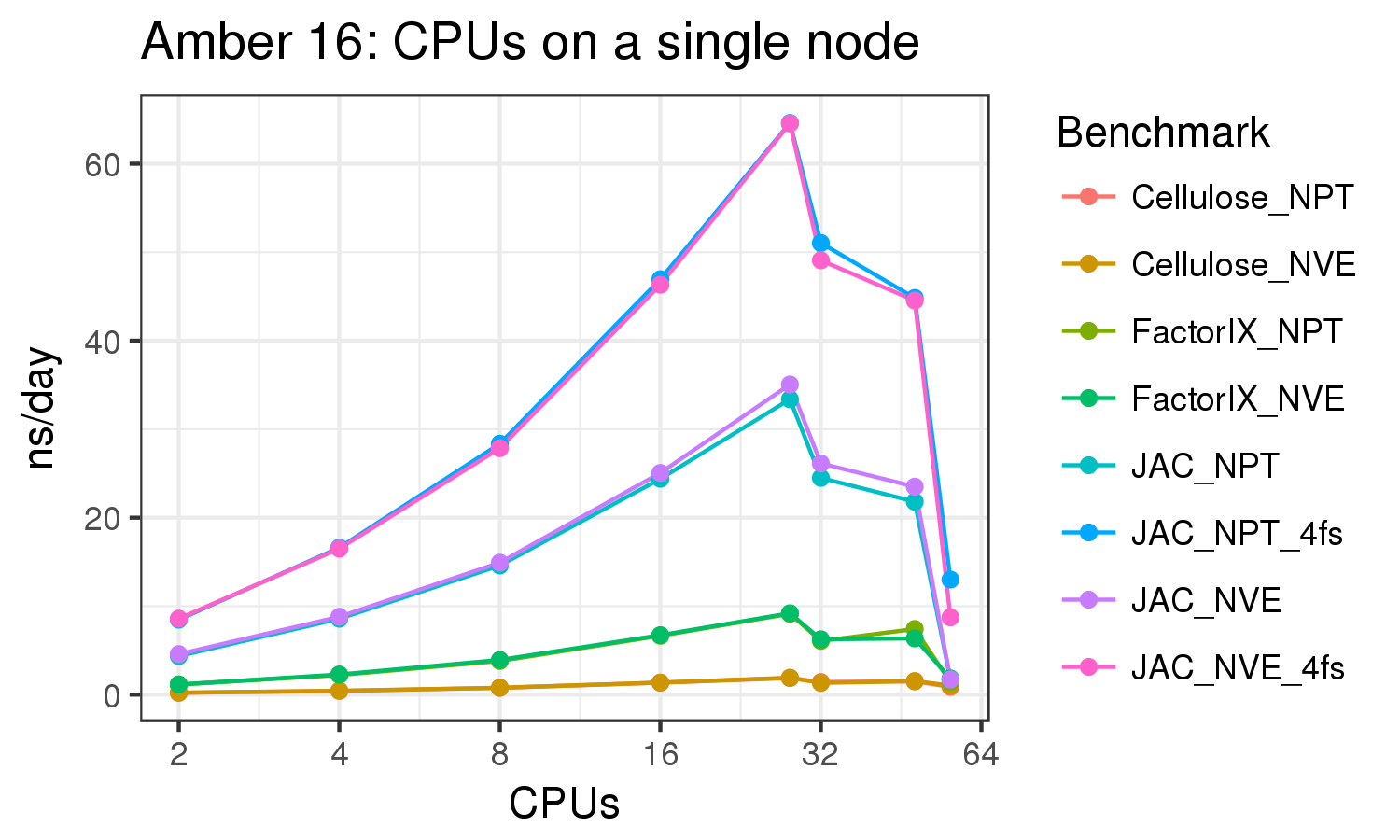
Amber 16 Benchmark suite, downloadable from here.
(The older Amber 14 benchmarks on Biowulf are also available).
Hardware:
CPU runs: 16x Intel Xeon E5-2650 v2 @2.6 GHz.
GPU runs: Nvidia driver version 352.39. (Note: driver version 367.48 gave very similar results).
Amber 16 with all patches as of 25 Dec 2016. Built with Intel 2015.1.133 compilers, CUDA 7.5, OpenMPI 2.0.1.
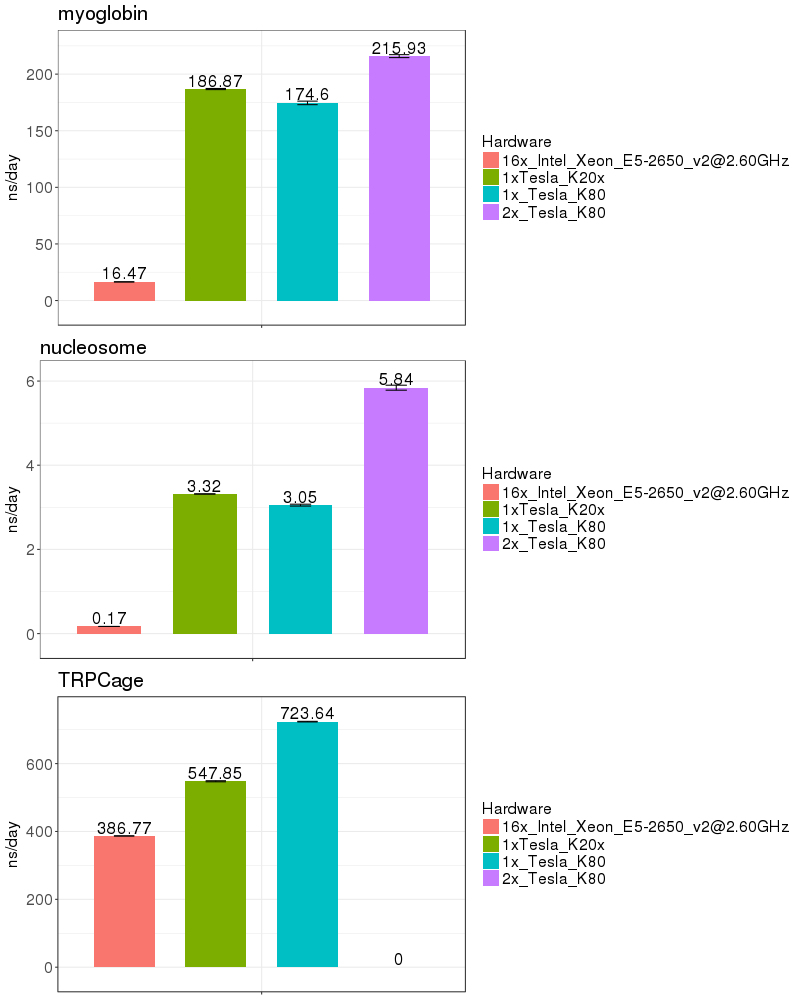
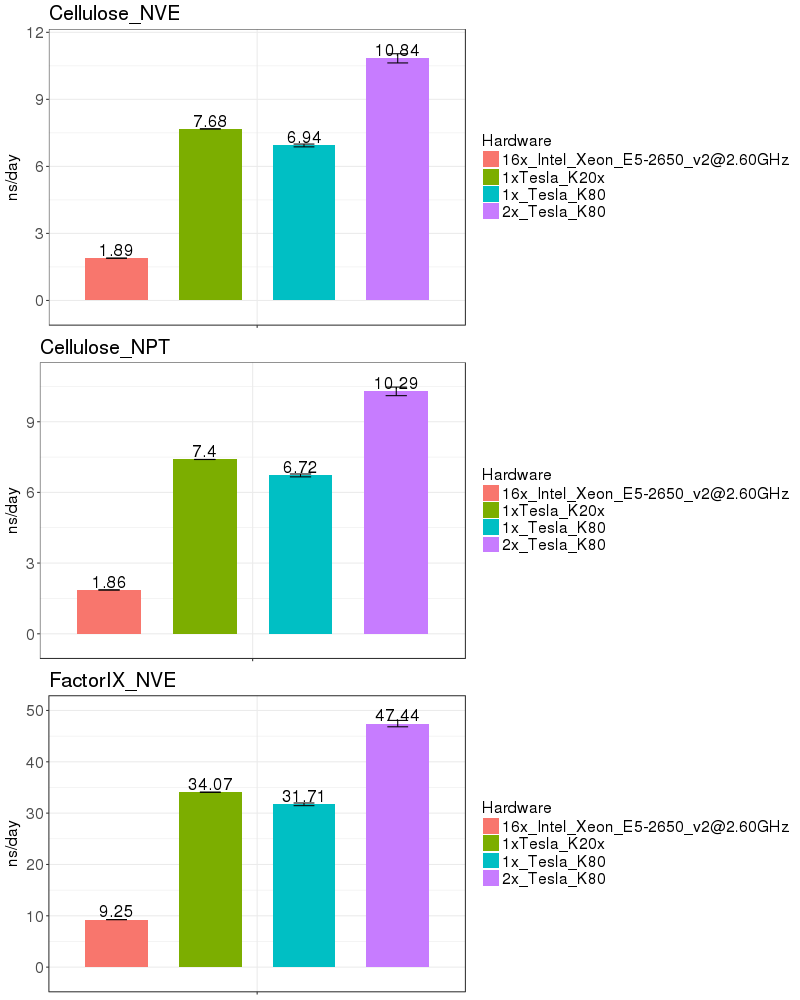
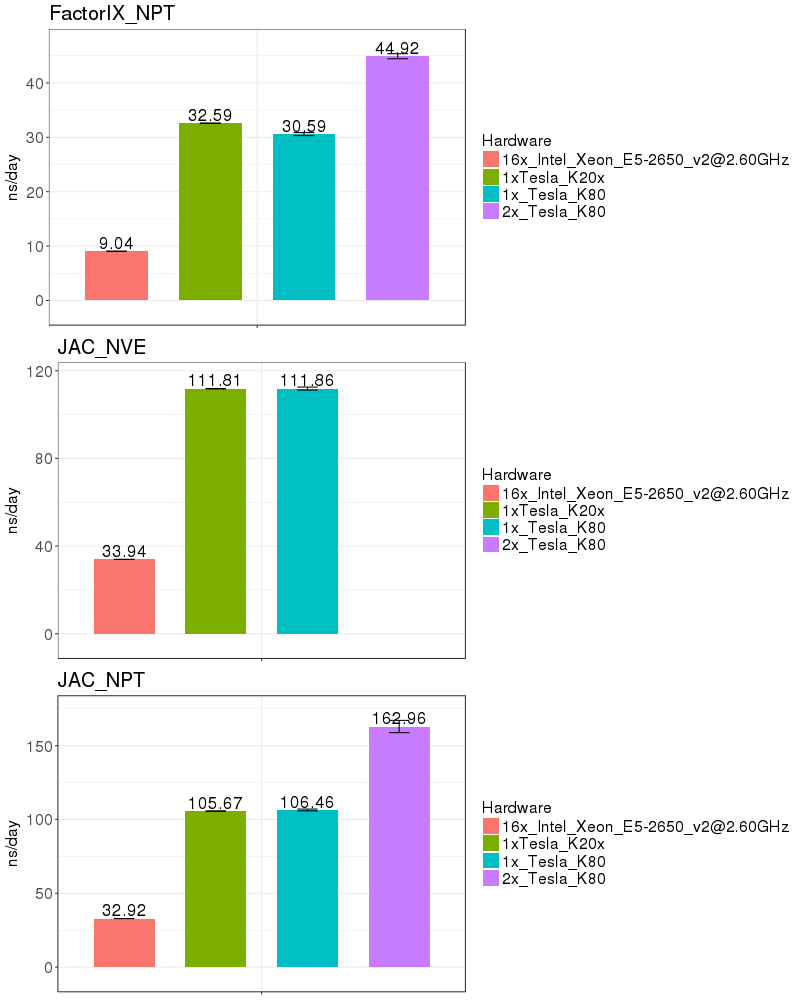
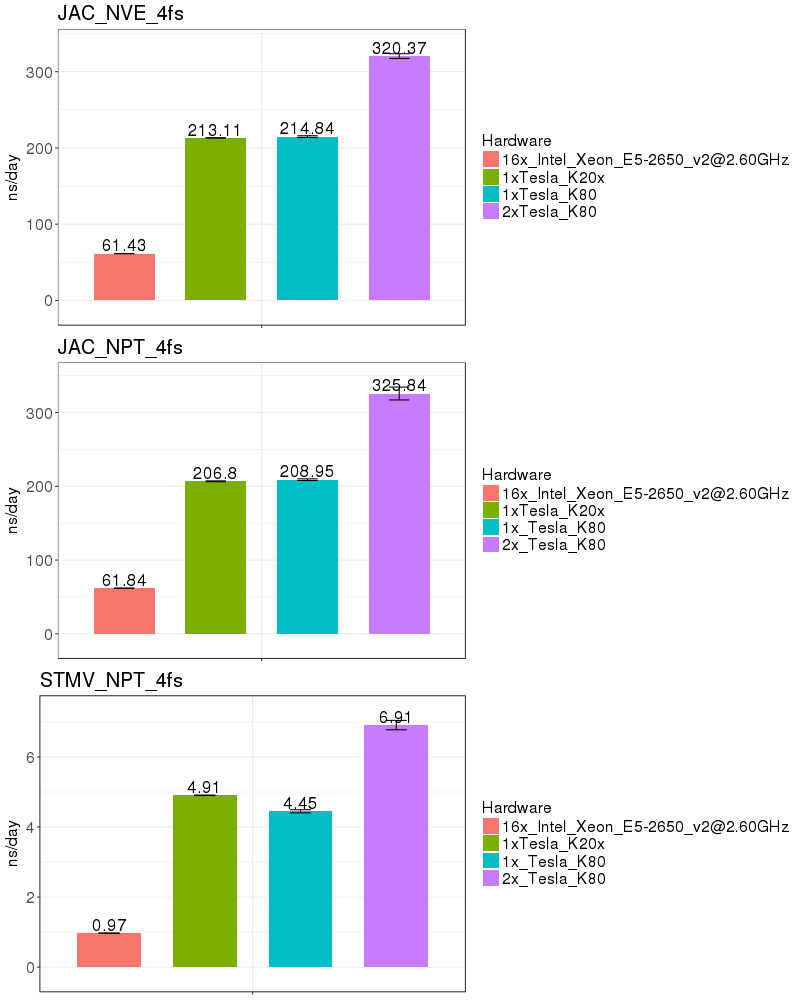
Based on these benchmarks, it is highly recomended that you run Amber on a GPU node. The performance on a single K20x and a single K80 are similar for most of the benchmarks run. It is not possible to run a single Amber job on both the K20s on a node, since those 2 GPUs do not have peer-to-peer communication. (see the Amber on GPUs page for an explanation of peer-to-peer communication).
If you run on the Biowulf K80 GPUs, it is possible to utilize 2 GPUs on a node, providing a performance advantage of about 50%. As of Jan 2017, the K80s on the system are funded by NCI_CCR and therefore restricted to CCR users. In March 2017, 72 additional K80s will be added to the Biowulf cluster and available to all users.

| Implicit Solvent (GB) | |
| |
| Explicit Solvent (PME) | |
| |
| |
| |