ReLeaSE (Reinforcement Learning for Structural Evolution) is an application for de-novo Drug Design
based on Reinforcement Learning. It integrates two deep neural networks: generative and
predictive, that are trained separately but are used jointly to generate novel targeted chemical libraries.
ReLeaSE uses simple representation of molecules by their simplified molecular input line entry specification (SMILES)
strings only.
This application, reimplemented in Keras from the original version developed in PyTorch,
is being used as a biological example by class #5 of the course
"Deep Learning by Example on Biowulf".
Allocate an interactive session and run the program. Sample session:
[user@biowulf]$ sinteractive --mem=160g --gres=gpu:k80,lscratch:10 -c8or, in order to use 4 GPUs,
[user@biowulf]$ sinteractive --mem=160g --gres=gpu:k80:4,lscratch:20 -c14 [user@cn4204 ~]$module load release [+] Loading cuDNN/7.6.5/CUDA-10.0 libraries... [+] Loading CUDA Toolkit 10.0.130 ... [+] Loading release 20200516The ReLeaSE application, as it is currently implemented on Biowulf, comprises the following source files:
[user@cn4204]$ ls $RELEASE_SRC data.py options.py predict.py visualize.py stackAugmentedRNN.py models.py train.py smiles.py utils.pyThe executables are:
[user@cn4204]$ ls $RELEASE_BIN predict.py train.py visualize.pyIn particular, the command line options of the executable train.py are as follows:
[user@cn4204]$ train.py -h
Using TensorFlow backend.
usage: train.py [-h] [-a ALPHA] [-b BATCH_SIZE] [-d DATA_TYPE] [-D]
[--dropout DROPOUT_FRACTION]
[--embedding_dim EMBEDDING_DIM] [--epochs NUM_EPOCHS]
[-f NUM_FOLDS] [-g num_gpus] [--delta DELTA]
[--hidden_size HIDDEN_SIZE] [-i input_checkpoint]
[--lr LR] -m TRAINING_MODE [--max_length MAX_LENGTH]
[--min_length MIN_LENGTH] [-n N_ROLLOUT]
[-N NUM_EPISODES] [-p PREDICT_LEN]
[-o output_checkpoint] [-O OPTIMIZER]
[-r RECURRENT_LAYER] [--stack_width STACK_WIDTH]
[--stack_depth STACK_DEPTH]
[--steps_per_epoch STEPS_PER_EPOCH] [-v]
optional arguments:
-h, --help show this help message and exit
-a ALPHA, --alpha ALPHA
reinforcement learning rate; default = 0.001
-b BATCH_SIZE, --batch_size BATCH_SIZE
input batch size; default=512
-d DATA_TYPE, --data_type DATA_TYPE
Data type: jak2 | logp; default = jak2
-D, --debug output debugging info
--dropout DROPOUT_FRACTION
the dropout fraction; default=0.2
--embedding_dim EMBEDDING_DIM
the dimension of the embedding space; default=512
--epochs NUM_EPOCHS, -e NUM_EPOCHS
number of iterations to train for; default = 51 for
jak2 and =25 for logp
-f NUM_FOLDS, --num_folds NUM_FOLDS
number of folds in cross-validation split; default=5
-g num_gpus, --num_gpus num_gpus
number of gpus to use; default=1
--delta DELTA discount rate; default = 0.98
--hidden_size HIDDEN_SIZE
hidden_size; default = 256
-i input_checkpoint, --input_checkpoint input_checkpoint
The name of the input checkpoint file; default = None
--lr LR learning rate (for generator) default: 3.e-4
--max_length MAX_LENGTH
max length of a SMILES string to be used for training;
default=None
--min_length MIN_LENGTH
min length of a SMILES string to be used for training;
default=None
-n N_ROLLOUT, --n_rollout N_ROLLOUT
number of rollout trajectories to average over;
default=16
-N NUM_EPISODES, --num_episodes NUM_EPISODES
number of episodesin the REINFORCE algorithm;
default=100
-p PREDICT_LEN, --predict_len PREDICT_LEN
max length of a sequence used to predict next
character; default=120
-o output_checkpoint, --output_checkpoint output_checkpoint
The name of the output checkpoint file; default=None
-O OPTIMIZER, --optimizer OPTIMIZER
Optimizer to use for training: adadelta | adam |
rmsprop; default=adam
-r RECURRENT_LAYER, --recurrent_layer RECURRENT_LAYER
Recurrent layer for generator: SimpleRNN | GRU | LSTM
| SA_SimpleRNN | SA_LSTM | SA_GRU (default)
--stack_width STACK_WIDTH
stack_width; default = 1
--stack_depth STACK_DEPTH
stack_depth; default = 100
--steps_per_epoch STEPS_PER_EPOCH
stack_width; default = None
-v, --verbose increase the verbosity level of output
required arguments:
-m TRAINING_MODE, --training_mode TRAINING_MODE
Training mode: generator | predictor | reinforce
To perform training of the predictor network using this executable, copy training data to the current folder:
[user@cn4204]$ cp -r $RELEASE_DATA/* .ReLeaSE takes as input data in the SMILES (Simplified Molecular Input Line Entry Specification) format. For example, the data used for training Generator model look like:
[user@cn4204]$ more data/chembl_22_clean_1576904_sorted_std_final.smi CCO CHEMBL545 C CHEMBL17564 CO CHEMBL14688 NCCS CHEMBL602 NCCN CHEMBL816 CN CHEMBL43280 C=O CHEMBL1255 CCN CHEMBL14449 ... CCC[N+](C)(C)CCO CHEMBL105059 CCCCCCCCN1CCCCC1 CHEMBL105218 ClCC(=O)c1ccccc1 CHEMBL105712 CCCCCCCNc1ccccc1 CHEMBL106128 ... CCOc1ccc(CN2CCN(Cc3ccon3)CC2CCO)cc1 CHEMBL1890577 Fc1ccc(Cc2nnc(o2)C(=O)NCC2CCCO2)cc1 CHEMBL1889842 COc1ccc(CC(N)c2csc(Nc3ccccn3)n2)cc1 CHEMBL1884880 CN1CCN(CC1)NC(=O)c1cccc(c1)C(F)(F)F CHEMBL1899891 O=C(NC1CCCCC1)C1CCCN1C(=O)Nc1ccccc1 CHEMBL1899656 CCc1nnc(NC(=O)CSc2nccn2Cc2ccccc2)s1 CHEMBL1901068 CCOC(=O)C(C)N1C=C(O)N(C1=O)c1ccccc1 CHEMBL1890667 ...A sample command to train Generator model on these data would be:
[user@cn4204]$ train.py -m generator -r SA_GRU -g 4 -b 1000 --lr 3.e-4 Using TensorFlow backend. ... Model: "sequential_1" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= embedding_1 (Embedding) (None, 11, 512) 45568 _________________________________________________________________ stack_augmented_rnn_1 (Stack (None, 11, 256) 617216 _________________________________________________________________ dropout_1 (Dropout) (None, 11, 256) 0 _________________________________________________________________ stack_augmented_rnn_2 (Stack (None, 11, 256) 420608 _________________________________________________________________ dropout_2 (Dropout) (None, 11, 256) 0 _________________________________________________________________ stack_augmented_rnn_3 (Stack (None, 256) 420608 _________________________________________________________________ dropout_3 (Dropout) (None, 256) 0 _________________________________________________________________ dense_1 (Dense) (None, 89) 22873 ================================================================= Total params: 1,526,873 Trainable params: 1,526,873 Non-trainable params: 0 _________________________________________________________________ Model compiled with loss='categorical_crossentropy', lr= 0.0003 Training generator ... ...or, using 4 GPUs,
[user@cn4204]$ train.py -m generator -r SA_GRU -g 4 -b 1000 --lr 3.e-4 ...The data for training Predictor model look as follows:
[user@cn4204]$ more data/jak2_data.csv SMILES,pIC50 O=S(=O)(Nc1cccc(-c2cnc3ccccc3n2)c1)c1cccs1,4.26 O=c1cc(-c2nc(-c3ccc(-c4cn(CCP(=O)(O)O)nn4)cc3)[nH]c2-c2ccc(F)cc2)cc[nH]1,4.34 NC(=O)c1ccc2c(c1)nc(C1CCC(O)CC1)n2CCCO,4.53 NCCCn1c(C2CCNCC2)nc2cc(C(N)=O)ccc21,4.56 CNC(=S)Nc1cccc(-c2cnc3ccccc3n2)c1,4.59 O=C(Nc1cccc(-c2cnc3ccccc3n2)c1)C1CC1,4.6 O=C(Nc1cccc(-c2cnc3ccccc3n2)c1)c1ccco1,4.61 COc1cc(Nc2nccc(-c3ccc(Cl)cc3)n2)cc(OC)c1OC,4.67 CN1C(=O)COc2c1cnc1ccc(Sc3nnc4c(F)cc(-c5cnn(C)c5)cn34)cc21,4.68 COc1ccc2c(c1)C(=O)N(c1nc(C(=O)Nc3cnccc3N3CCNCC3)cs1)C2,4.68 ...- for predicting logP property value:
[user@cn4204]$ more data/logp_labels.csv ,SMILES,Kow 1,CC1CC2C3CCC4=CC(=O)C=CC4(C)C3(F)C(O)CC2(C)C1(O)C(=O)CO,1.885 2,CC(=O)OCC(=O)C1(O)CCC2C3CCC4=CC(=O)CCC4(C)C3C(O)CC21C,2.19 3,CC(=O)OCC(=O)C1(O)CCC2C3CCC4=CC(=O)CCC4(C)C3C(=O)CC21C,2.1 4,CCC1(c2ccccc2)C(=O)NC(=O)NC1=O,1.47 5,COC12C(COC(N)=O)C3=C(C(=O)C(C)=C(N)C3=O)N1CC1NC12,-0.4 6,CCC1(CC)C(=O)NC(=O)N(C)C1=O,1.15 7,CCC1(c2ccccc2)NC(=O)N(C)C1=O,1.69 8,O=P1(N(CCCl)CCCl)NCCCO1,0.63 9,CC(O)C(=O)O,-0.72 10,CC12CCC(=O)C=C1CCC1C2C(O)CC2(C)C(C(=O)CO)CCC12,1.94 11,CC12CCC(=O)C=C1CCC1C2C(O)CC2(C)C1CCC2(O)C(=O)CO,1.61 12,CC12C=CC(=O)C=C1CCC1C2C(O)CC2(C)C1CCC2(O)C(=O)CO,1.62 13,CC12CCC3c4ccc(O)cc4CCC3C1CC(O)C2O,2.45 14,CC12CCC3c4ccc(O)cc4CCC3C1CCC2O,4.01 15,Clc1ccc(C(c2ccc(Cl)cc2)C(Cl)(Cl)Cl)cc1,6.91 16,O=C(O)c1c(Cl)cccc1Cl,2.23 17,c1ccc2c(c1)cc1ccc3cccc4ccc2c1c34,6.13 18,CCCCC1C(=O)N(c2ccccc2)N(c2ccccc2)C1=O,3.16 ...Sample commands to train the Predictor model are:
[user@cn4204]$ train.py -d jak2 -m predictor -g 1 -b 128 --lr 0.0001 -e 500 ... Model: "sequential_1" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= embedding_1 (Embedding) (None, 120, 512) 45568 _________________________________________________________________ lstm_1 (LSTM) (None, 128) 328192 _________________________________________________________________ dropout_1 (Dropout) (None, 128) 0 _________________________________________________________________ batch_normalization_1 (Batch (None, 128) 512 _________________________________________________________________ dense_1 (Dense) (None, 128) 16512 _________________________________________________________________ activation_1 (Activation) (None, 128) 0 _________________________________________________________________ dense_2 (Dense) (None, 1) 129 _________________________________________________________________ activation_2 (Activation) (None, 1) 0 ================================================================= Total params: 390,913 Trainable params: 390,657 Non-trainable params: 256 _________________________________________________________________ Reading predictor data ... ... [user@cn4204]$ train.py -d logp -m predictor -g 1 -b 128 --lr 0.0001 -e 500 ... [user@cn4204]$ train.py -m reinforce -d jak2 ... [user@cn4204]$ train.py -m reinforce -d jak2 -i checkpoints/generator.weights.SA_GRU.1.h5,checkpoints/predictor.weights.jak2.h5 ...After training the models, the checkpoints (model weights or the entire model) are save in the checkpoints folder.
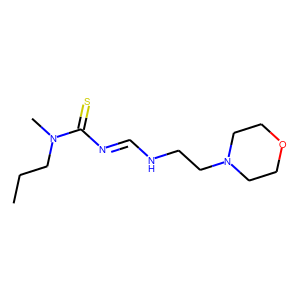
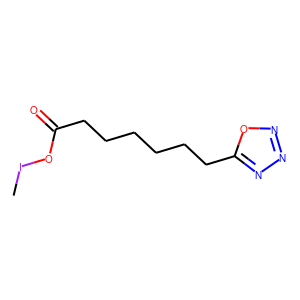
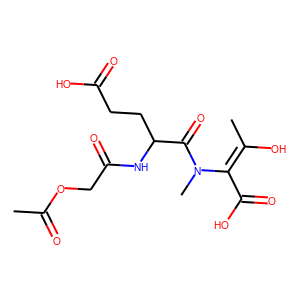
[user@cn4204]$ predict.py -i checkpoints/generator.weights.SA_GRU.1.h5 ... generated SMILES = C(NCCN2CCOCC2)=NC(=S)N(CCC)C [user@cn4204]$ predict.py -i checkpoints/generator.weights.SA_GRU.2.h5 --stack_width 2 ... Generated SMILES= 'CIOC(=O)CCCCCCc1nnno1' [user@cn4204]$ predict.py -r LSTM -i checkpoints/generator.weights.LSTM.h5 ... Generated SMILES= 'CC(O)=C(C(O)=O)N(C)C(=O)C(CCC(O)=O)NC(=O)COC(C)=O' [user@cn4204]$ predict.py -i checkpoints/reinforce.weights.jak2.SA_GRU.1.h5,checkpoints/predictor.weights.jak2.h5 ... Generated SMILES= 'C[CH2]C=C(C)CCSSSCCCN=C=N' jak2 property value= 0.4330919 ... [user@cn4204]$ predict.py -s 'CC(Cl)=Cc3ccc(Cl)cc3' -i checkpoints/predictor.model.jak2.h5 ... Input SMILES= 'CC(Cl)=Cc3ccc(Cl)cc3' jak2 property value= 0.03461966NOTE: since each the SMILES token is sampled randomly from the respective distribution, different runs of the same command may produce different SMILES strings.
[user@cn4204]$ visualize.py -s "C(NCCN2CCOCC2)=NC(=S)N(CCC)C"
[user@cn4204]$ visualize.py -s "CIOC(=O)CCCCCCc1nnno1"
[user@cn4204]$ visualize.py -s "CC(O)=C(C(O)=O)N(C)C(=O)C(CCC(O)=O)NC(=O)COC(C)=O"
[user@cn4204 ~]$ exit salloc.exe: Relinquishing job allocation 46116226 [user@biowulf ~]$