The DeepEMhancer is a python package designed to perform post-processing of cryo-EM map.
deepemhancer -h
cp -r /usr/local/apps/DeepEMhancer/0.13/deepEMhancerModels/ /data/$USER/
tensorflow/2 instead of tensorflow/1. Please contact staff@hpc.nih.gov if you need to use the older version. Please copy the deep learning models for tf2 to avoid errors.
cp -r /usr/local/apps/DeepEMhancer/0.13/deepEMhancerModels/ /data/$USER/
Allocate an interactive session and run the program.
Sample session (user input in bold):
[user@biowulf]$ sinteractive --gres=gpu:p100:1 --mem=8g
salloc.exe: Pending job allocation 46116226
salloc.exe: job 46116226 queued and waiting for resources
salloc.exe: job 46116226 has been allocated resources
salloc.exe: Granted job allocation 46116226
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3144 are ready for job
[user@cn3144 ~]$ module load DeepEMhancer
[user@cn3144 ~]$ mkdir /data/$USER/DeepEMhancer_test/
[user@cn3144 ~]$ cd /data/$USER/DeepEMhancer_test/
[user@cn3144 ~]$ deepemhancer -h
usage: deepemhancer -i INPUTMAP -o OUTPUTMAP
[-p {wideTarget,tightTarget,highRes}] [-i2 HALFMAP2]
[-s SAMPLINGRATE] [--noiseStats NOISE_MEAN NOISE_STD]
[-m BINARYMASK]
[--deepLearningModelPath PATH_TO_MODELS_DIR]
[--cleaningStrengh CLEANINGSTRENGH] [-g GPUIDS]
[-b BATCH_SIZE] [-h] [--download [DOWNLOAD_DEST]]
DeepEMHancer. Deep post-processing of cryo-EM maps. https://github.com/rsanchezgarc/deepEMhancer
optional arguments:
-h, --help show this help message and exit
--download [DOWNLOAD_DEST]
download default DeepEMhancer models. They will be
saved at /home/$USER/.local/share/deepEMhancerModels/pr
oduction_checkpoints if no path provided
Main options:
-i INPUTMAP, --inputMap INPUTMAP
Input map to process or half map number 1. This map
should be unmasked and not sharpened (Do not use post-
processed maps, only maps directly obtained from
refinement). If half map 1 used, do not forget to also
provide the half map 2 using -i2
-o OUTPUTMAP, --outputMap OUTPUTMAP
Output fname where post-processed map will be saved
-p {wideTarget,tightTarget,highRes}, --processingType {wideTarget,tightTarget,highRes}
Select the deep learning model you want to use.
WideTarget will produce less sharp results than
tightTarget. HighRes is only recommended for overal
FSC resolution < 4 A This option is igonred if
normalization mode 2 is selected
-i2 HALFMAP2, --halfMap2 HALFMAP2
(Optional) Input half map 2 to process
-s SAMPLINGRATE, --samplingRate SAMPLINGRATE
(Optional) Sampling rate (A/voxel) of the input map.
If not provided, the sampling rate will be read from
mrc file header
Normalization options (auto normalization is applied if no option selected):
--noiseStats NOISE_MEAN NOISE_STD
(Optional) Normalization mode 1: The statisitcs of the
noise to normalize (mean and standard deviation) the
input. Preferred over binaryMask but ignored if
binaryMask provided. If not --noiseStats nor
--binaryMask provided, nomralization params will be
automatically estimated, although, in some rare cases,
estimation may fail or be less accurate
-m BINARYMASK, --binaryMask BINARYMASK
(Optional) Normalization mode 2: A binaryMask (1
protein, 0 no protein) used to normalize the input. If
no normalization mode provided, automatic
normalization will be carried out. Supresses
--precomputedModel option
Alternative options:
--deepLearningModelPath PATH_TO_MODELS_DIR
(Optional) Directory where a non default deep learning
model is located (model is selected using
--precomputedModel) or a path to hd5 file containing
the model
--cleaningStrengh CLEANINGSTRENGH
(Optional) Post-processing step to remove small
connected components (hide dust). Max relative size of
connected components to remove 0<s<1 or -1 to
deactivate. Default: -1
Computing devices options:
-g GPUIDS, --gpuIds GPUIDS
The gpu(s) where the program will be executed. If more
that 1, comma seppared. E.g -g 1,2,3. Set to -1 to use
only cpu (very slow). Default: 0
-b BATCH_SIZE, --batch_size BATCH_SIZE
Number of cubes to process simultaneously. Lower it if
CUDA Out Of Memory error happens and increase it if
low GPU performance observed. Default: 8
examples:
+ Download deep learning models
deepemhancer --download
+ Post-process input map path/to/inputVol.mrc and save it at path/to/outputVol.mrc using default deep model tightTarget
deepemhancer -i path/to/inputVol.mrc -o path/to/outputVol.mrc
+ Post-process input map path/to/inputVol.mrc and save it at path/to/outputVol.mrc using high resolution deep model
deepemhancer -p highRes -i path/to/inputVol.mrc -o path/to/outputVol.mrc
+ Post-process input map path/to/inputVol.mrc and save it at path/to/outputVol.mrc using a deep learning model located in path/to/deep/learningModel
deepemhancer -c path/to/deep/learningModel -i path/to/inputVol.mrc -o path/to/outputVol.mrc
+ Post-process input map path/to/inputVol.mrc and save it at path/to/outputVol.mrc using high resolution deep model and providing normalization information (mean
and standard deviation of the noise)
deepemhancer -p highRes -i path/to/inputVol.mrc -o path/to/outputVol.mrc --noiseStats 0.12 0.03
[user@cn3144 ~]$ deepemhancer --deepLearningModelPath /data/$USER/deepEMhancerModels/production_checkpoints -i input.mrc -o output.mrc
[user@cn3144 ~]$ exit
salloc.exe: Relinquishing job allocation 46116226
[user@biowulf ~]$
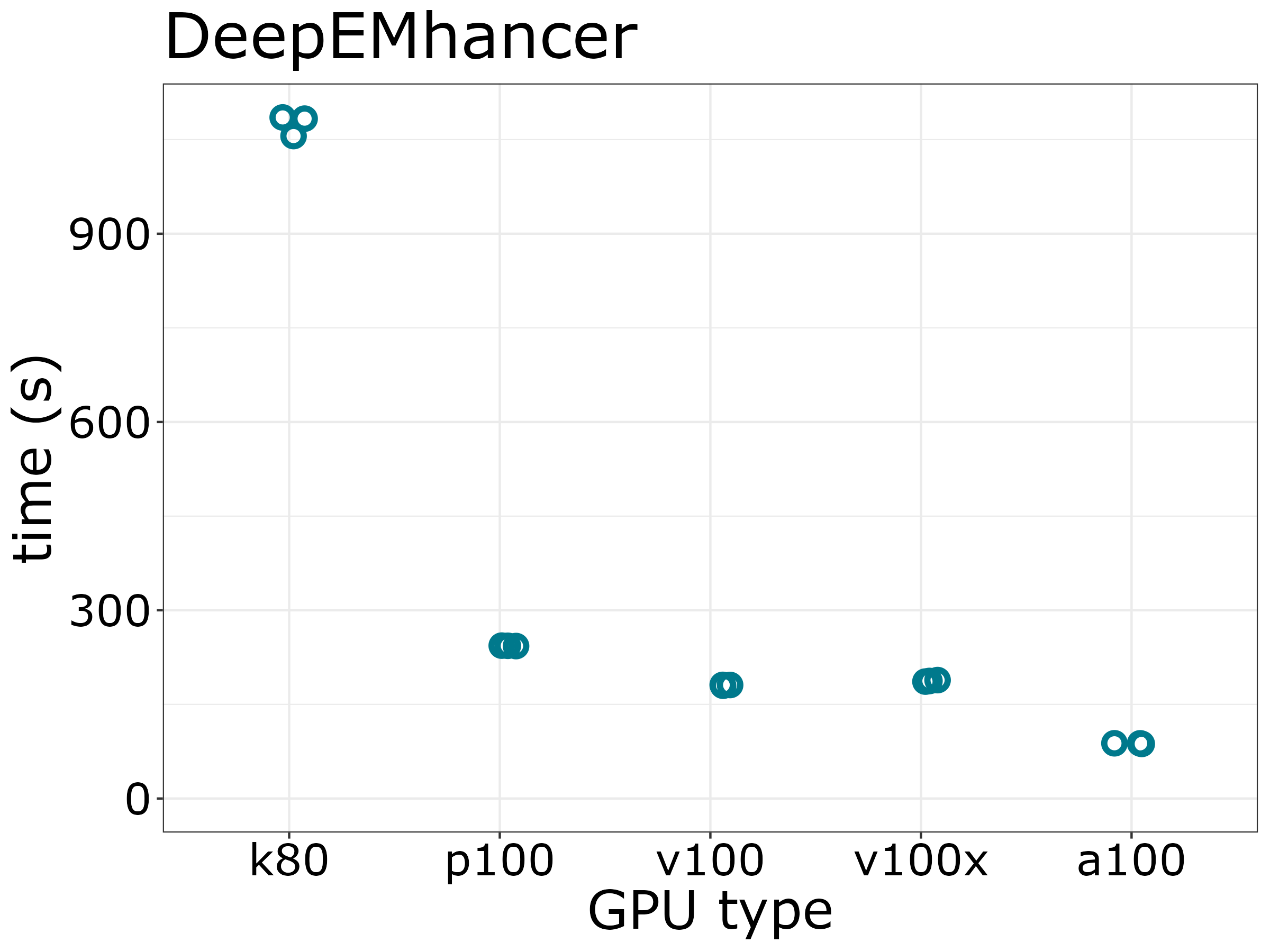
To estimate the runtimes of DeepEMhancer on CentOS7, we run DeepEMhancer on 4 types of GPUs which 4G memory, 2 CPUs and 1 GPU (3 replicates). K80 GPU took about 3 times longer as other GPUs. p100 GPU is about as good as other modern GPUs.

Create a batch input file (e.g. DeepEMhancer.sh). For example:
#!/bin/bash
set -e
module load DeepEMhancer
# -g will assign GPUIDs, always start with 0 for batch job
deepemhancer --deepLearningModelPath /data/$USER/deepEMhancerModels/production_checkpoints -i input_half1.mrc -i2 input_half2.mrc -o output.mrc -g 0
Submit this job using the Slurm sbatch command.
sbatch --partition=gpu --gres=gpu:p100:1 --mem=8g DeepEMhancer.sh
Create a swarmfile (e.g. DeepEMhancer.swarm). For example:
cd dir1;deepemhancer --deepLearningModelPath /data/$USER/deepEMhancerModels/production_checkpoints -i input1.mrc -o output1.mrc cd dir1;deepemhancer --deepLearningModelPath /data/$USER/deepEMhancerModels/production_checkpoints -i input2.mrc -o output2.mrc
Submit this job using the swarm command.
swarm -f DeepEMhancer.swarm [-t #] [-g #] --partition=gpu --gres=gpu:p100:1 --module DeepEMhancerwhere
| -g # | Number of Gigabytes of memory required for each process (1 line in the swarm command file) |
| -t # | Number of threads/CPUs required for each process (1 line in the swarm command file). |
| --module DeepEMhancer | Loads the DeepEMhancer module for each subjob in the swarm |