Amber18 benchmarks
Amber 16 Benchmark suite, downloadable from here.
(The older Amber 16 and Amber 14 benchmarks on Biowulf are also available).
Hardware:
- CPU runs: 28 CPUs, Intel(R)_Xeon(R)_CPU_E5-2680_v4_@_2.40GHz
- Intel(R)_Xeon(R)_CPU_E5-2680_v4_@_2.40GHz, Tesla K80 -- benchmark runs with 1 GPU, 2 GPU, 4 GPUs
- Intel(R)_Xeon(R)_CPU_E5-2680_v4_@_2.40GHz, Tesla_P100-PCIE-16GB -- benchmark runs with 1 GPU, 2 GPU, 4 GPUs
- Intel(R)_Xeon(R)_CPU_E5-2680_v4_@_2.40GHz, Tesla_V100-PCIE-16GB -- benchmark runs with 1 GPU, 2 GPU, 4 GPUs
Amber 18 with all patches as of 3 Oct 2018. Built with Intel 2015.1.133 compilers, CUDA 9.2, OpenMPI 2.1.2.
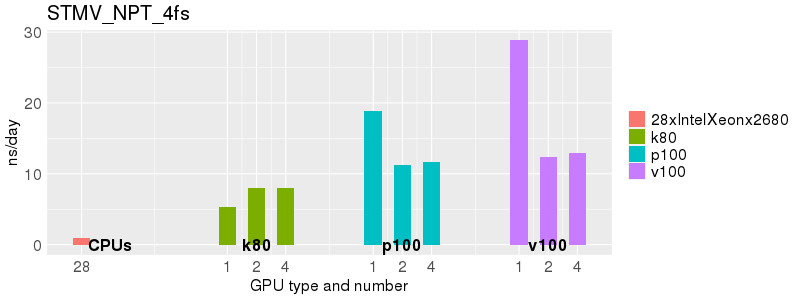
Based on these benchmarks, there is a significant performance advantage to running Amber on the GPU nodes, especially the P100s and V100s, rather than on a CPU-only node. However, there is little advantage to using more than 1 GPU.
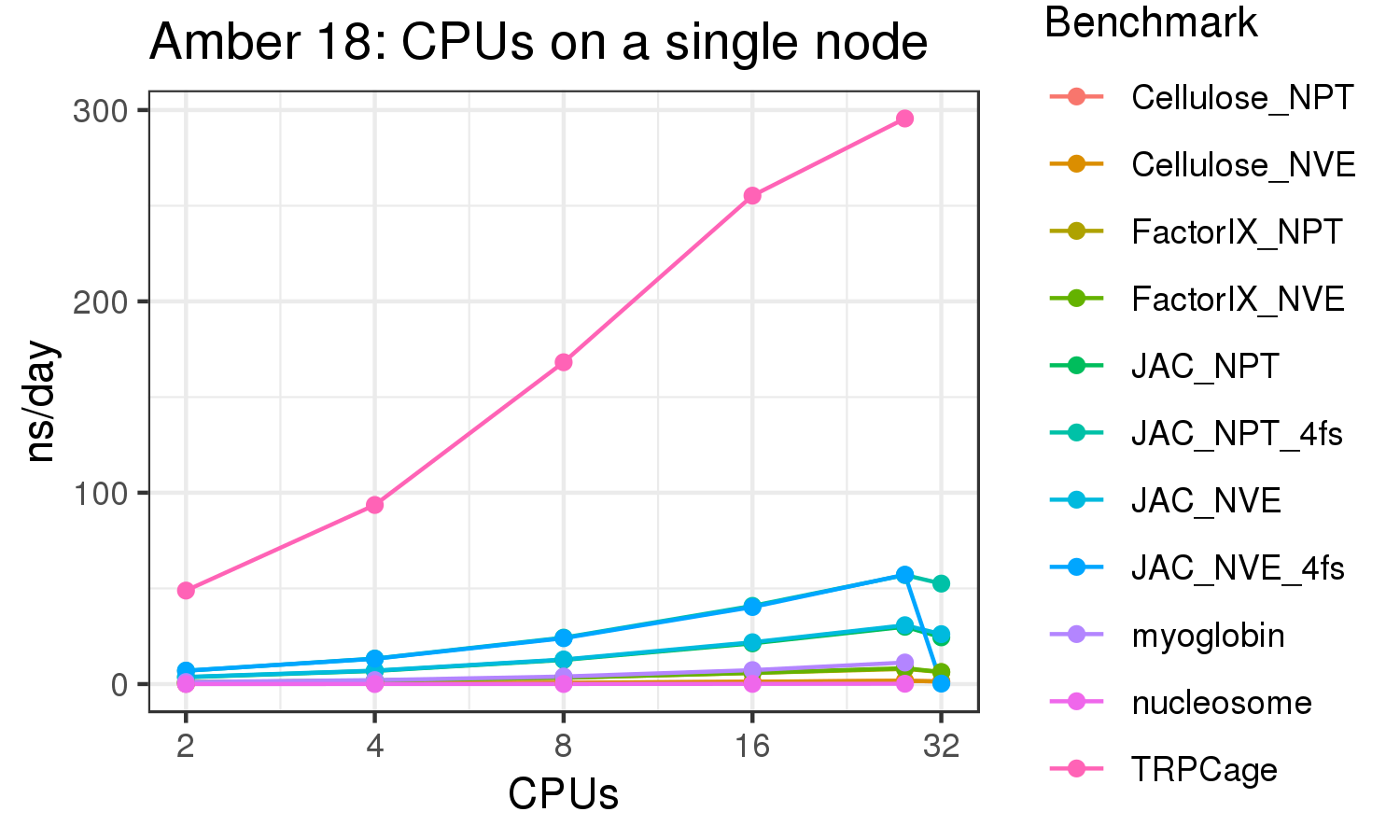
The benchmark above was run on a Intel Xeon E5-2680 v4 @2.5GHz with 28 cores (56 hyperthreaded cores). As with other MD applications, the performance drops when more than 28 MPI processes (i.e. 1 process per physical core) are run. Thus, if running on CPUs, it is important to use the '--ntasks-per-core=1' flag when submitting the job, to ensure that only 1 MPI process is run on each physical core. If it is possible to run on a GPU node, you will get significantly better performance as shown in the benchmarks below.
Implicit Solvent (GB)
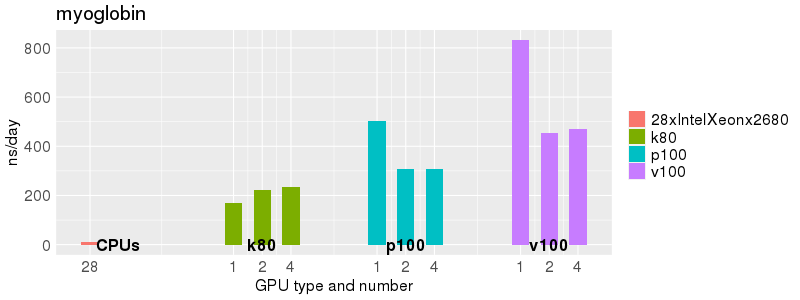
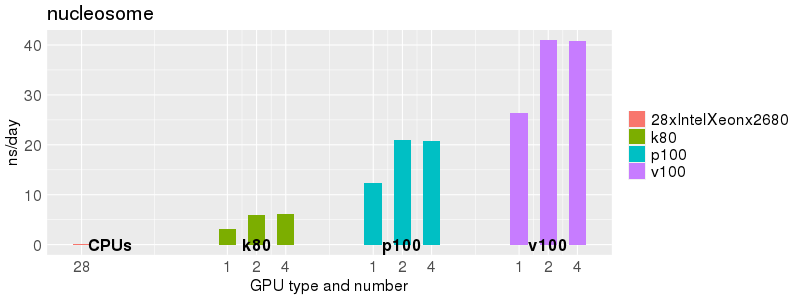
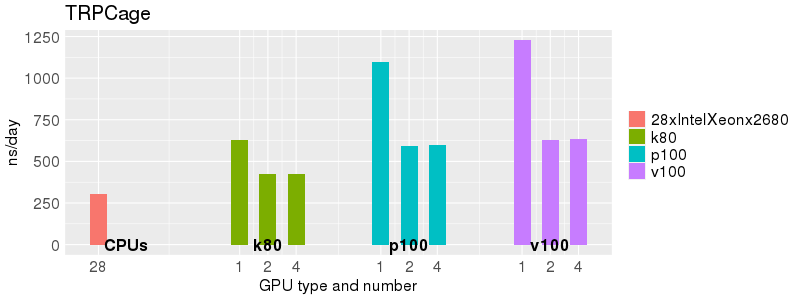 Explicit Solvent (PME)
Explicit Solvent (PME)
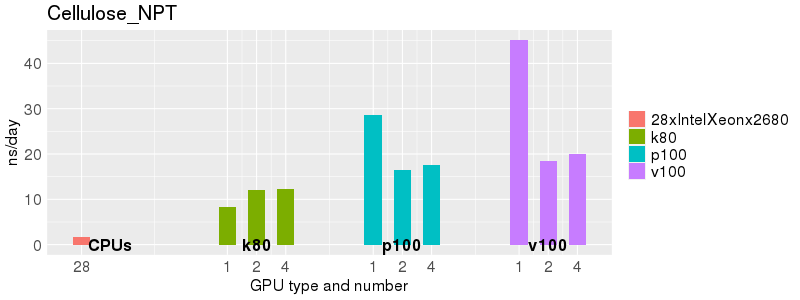
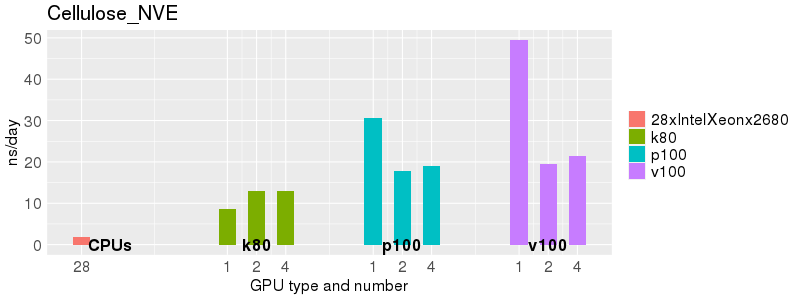
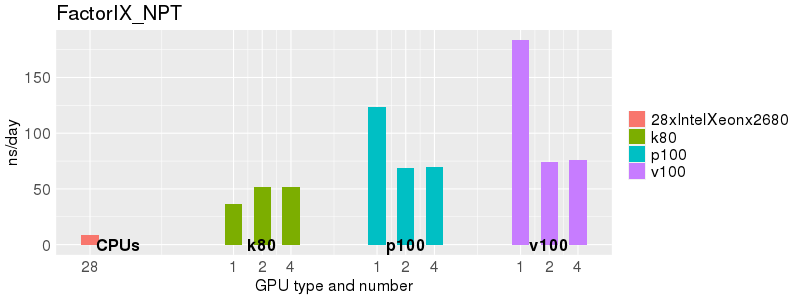
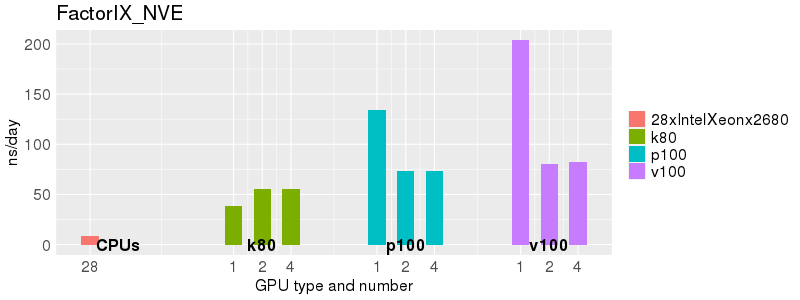
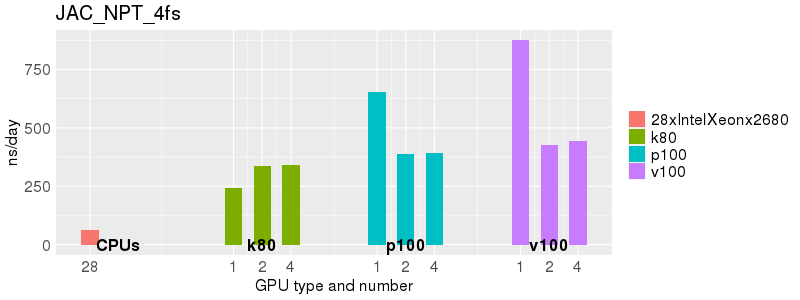
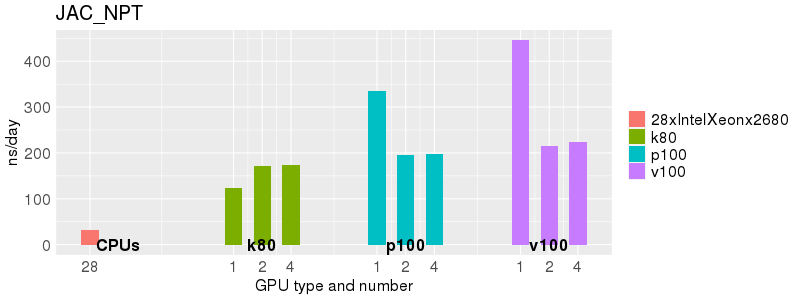
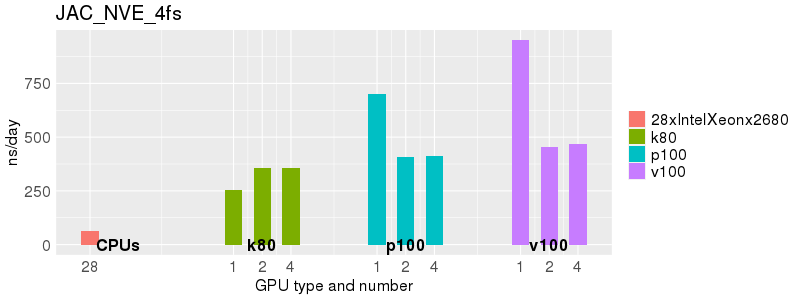
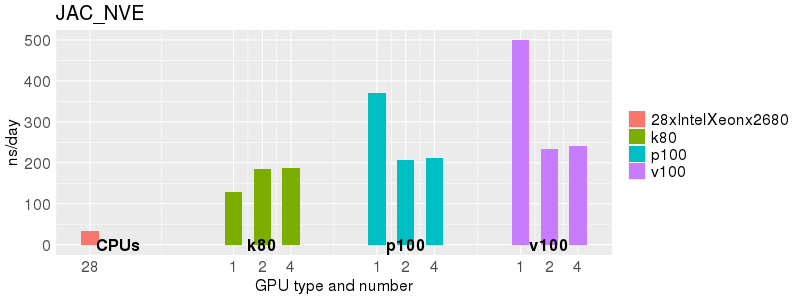




 Explicit Solvent (PME)
Explicit Solvent (PME)








