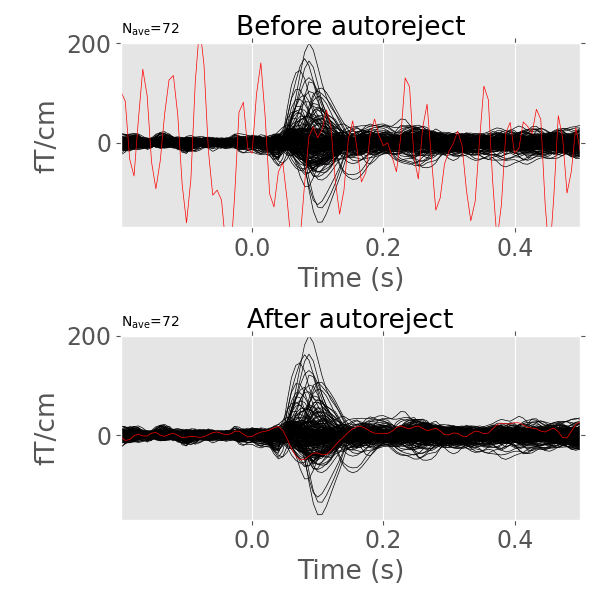
This is a library to automatically reject bad trials and repair bad sensors in magneto-/electroencephalography (M/EEG) data.

autoreject is installed as a singularity container, which means it came with it's own python environment.
It could be run as command line or through jupyter notebook.
Allocate an interactive session and run the program.
Sample session (user input in bold):
[user@biowulf]$ sinteractive --cpus-per-task=10 --mem=10G --gres=lscratch:200 --tunnel
salloc.exe: Pending job allocation 46116226
salloc.exe: job 46116226 queued and waiting for resources
salloc.exe: job 46116226 has been allocated resources
salloc.exe: Granted job allocation 46116226
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3144 are ready for job
Created 1 generic SSH tunnel(s) from this compute node to
biowulf for your use at port numbers defined
in the $PORTn ($PORT1, ...) environment variables.
Please create a SSH tunnel from your workstation to these ports on biowulf.
On Linux/MacOS, open a terminal and run:
ssh -L 33327:localhost:33327 biowulf.nih.gov
For Windows instructions, see https://hpc.nih.gov/docs/tunneling
[user@cn3144]$ module load autoreject
[user@cn3144]$ python -c "import autoreject"
[user@cn3144]$ cd /lscratch/$SLURM_JOB_ID
[user@cn3144]$ cp -r ${AUTOREJECT_TEST_DATA:-none}/mne_data .
[user@cn3144]$ jupyter notebook --ip localhost --port $PORT1 --no-browser
[I 17:11:40.505 NotebookApp] Serving notebooks from local directory
[I 17:11:40.505 NotebookApp] Jupyter Notebook 6.4.10 is running at:
[I 17:11:40.505 NotebookApp] http://localhost:37859/?token=xxxxxxxx
[I 17:11:40.506 NotebookApp] or http://127.0.0.1:37859/?token=xxxxxxx
[I 17:11:40.506 NotebookApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[C 17:11:40.512 NotebookApp]
To access the notebook, open this file in a browser:
file:///home/apptest1/.local/share/jupyter/runtime/nbserver-29841-open.html
Or copy and paste one of these URLs:
http://localhost:37859/?token=xxxxxxx
or http://127.0.0.1:37859/?token=xxxxxxx
Then you can open a browser from your computer to connect to the jupyter notebook:
[user@cn3144 ~]$ exit salloc.exe: Relinquishing job allocation 46116226 [user@biowulf ~]$
Create a batch input file (e.g. autoreject.sh). For example:
#!/bin/bash #SBATCH --cpus-per-task=10 #SBATCH --mem=10G #SBATCH --time=2:00:00 #SBATCH --partition=norm set -e module load autoreject # command
Submit the job:
sbatch autoreject.sh
Create a swarmfile (e.g. job.swarm). For example:
cmd1
cmd2
cmd3
Submit this job using the swarm command.
swarm -f job.swarm [-g #] --module autorejectwhere
| -g # | Number of Gigabytes of memory required for each process (1 line in the swarm command file) |
| --module | Loads the module for each subjob in the swarm |