
The High Performing Computation (HPC) group at the National Institutes of Health provides computational resources and support for the NIH intramural research community.
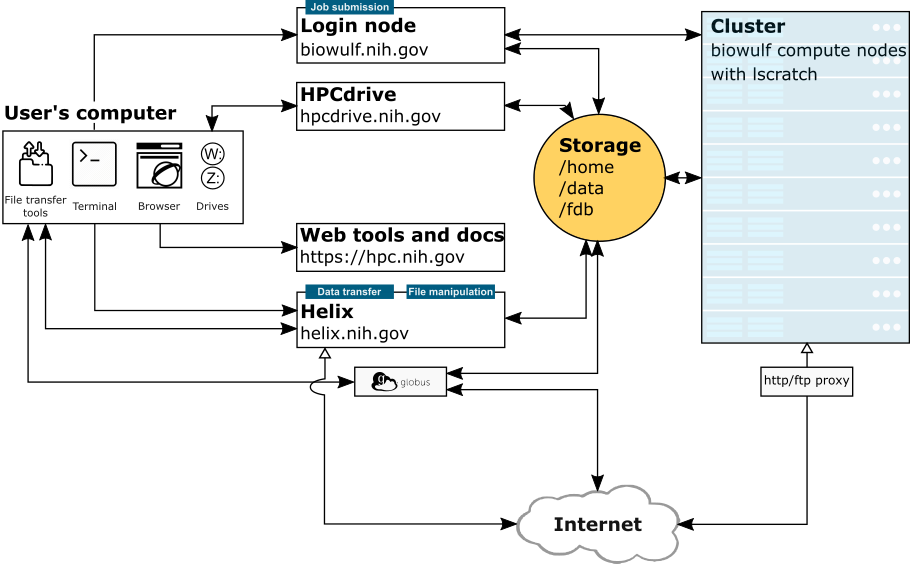
Helix (helix.nih.gov) is the interactive data transfer and file management node for the NIH HPC Systems. Users should run all such processes (scp, sftp, Aspera transfers, rsync, wget/curl, large file compressions, etc.) on this system. Scientific applications are not available on Helix. Helix is a 48 core (4 X 3.00 GHz 12-core Xeon™ Gold 6136) system with 1.5 TB of main memory running RedHat Enterprise Linux 8 and has a direct connection to the NIH network.
Each user on Helix is restricted to 6 CPUs and 32GB of memory. That means that any file transfer or compression processes should not use more than 6 threads total to run efficiently and avoid overloading the network and file systems. For example, an SRA download uses 6 threads by default, so a user should not run more than 1 such download at a time.
| Helix | Biowulf Login Node | Biowulf Cluster Compute Nodes | |
|---|---|---|---|
| Purpose | Dedicated data transfer system. No scientific programs can be run. | Submission of jobs. No scientific programs.. | Most computational processes, run via batch jobs or sinteractive sessions. |
| Network | direct connection to the NIH network (and internet) | connects to the NIH network (and internet) via proxy server | |
| System | Single system shared by all users | Single system shared by all users | 4000+ compute nodes with a total of 100,000+ compute cores. CPUs and memory for a job are dedicated to that job during its walltime and do not compete with other users. |
| /scratch vs /lscratch | /scratch is accessible | /scratch is accessible | /scratch is not accessible from the compute nodes. Each node has its own local disk (/lscratch) which can be allocated to a job (more info) |