Biowulf users with COVID-related projects should contact the HPC staff to get increased priority for their jobs.
|
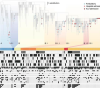
Covid-Related Research on Biowulf
0 Million CPU hours used
|
| ||
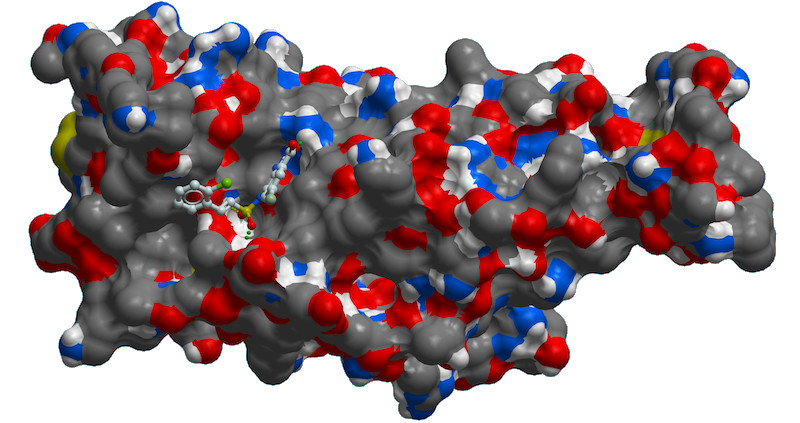
| Biowulf was used for screening of large virtual libraries containing billions of compounds
and allowed for identification of small molecules that bind to SARS-CoV2 spike protein and inhibit viral infection.
The image depicts one of such molecules bound to the receptor-binding domain of the protein. [Nadya Tarasova, NCI]
| 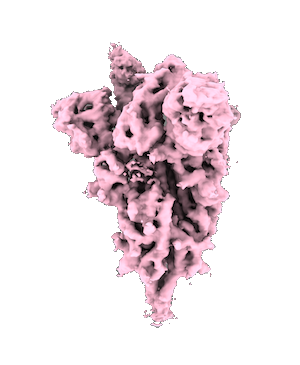
3D structural model | of the coronavirus spike protein [Mario Borgnia, NIEHS] |
-
Database is currently unavailable.
Biowulf users with COVID-related projects should contact the HPC staff to get increased priority for their jobs.
105 peer-reviewed papers
 Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer
Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer
McBride, DS; Garushyants, SK; Franks, J et al.
Nat Commun (2023)
 Exposure to lung-migrating helminth protects against murine SARS-CoV-2 infection through macrophage-dependent T cell activation
Exposure to lung-migrating helminth protects against murine SARS-CoV-2 infection through macrophage-dependent T cell activation
Oyesola, OO; Hilligan, KL; Namasivayam, S et al.
Sci Immunol (2023)
Optimized quantification of intra-host viral diversity in SARS-CoV-2 and influenza virus sequence data
Roder, AE; Johnson, KEE; Knoll, M et al.
mBio (2023)
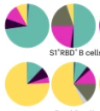 Adaptive immune responses to SARS-CoV-2 persist in the pharyngeal lymphoid tissue of children
Adaptive immune responses to SARS-CoV-2 persist in the pharyngeal lymphoid tissue of children
Xu, Q; Milanez-Almeida, P; Martins, AJ et al.
Nat Immunol (2023)
 Topoisomerase 3b is dispensable for replication of a positive-sense RNA virus--murine coronavirus
Topoisomerase 3b is dispensable for replication of a positive-sense RNA virus--murine coronavirus
Zhang, T; Su, S; Altouma, V et al.
Antiviral Res (2022)
 A conserved oligomerization domain in the disordered linker of coronavirus nucleocapsid proteins
A conserved oligomerization domain in the disordered linker of coronavirus nucleocapsid proteins
Zhao, H; Wu, D; Hassan, SA et al.
Sci Adv (2023)
 Wastewater surveillance uncovers regional diversity and dynamics of SARS-CoV-2 variants across nine states in the USA
Wastewater surveillance uncovers regional diversity and dynamics of SARS-CoV-2 variants across nine states in the USA
Fontenele, RS; Yang, Y; Driver, EM et al.
Sci Total Environ (2023)
 Identification of SARS-CoV-2 Mpro inhibitors containing P1’ 4-fluorobenzothiazole moiety highly active against SARS-CoV-2
Identification of SARS-CoV-2 Mpro inhibitors containing P1’ 4-fluorobenzothiazole moiety highly active against SARS-CoV-2
Higashi-Kuwata, N; Tsuji, K; Hayashi, H et al.
Nat Commun (2023)
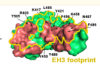 Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD
Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD
Chen, Y; Prévost, J; Ullah, I et al.
iScience (2023)
Analysis of immune responses in CLL patients after heterologous COVID-19 vaccination
Lee, HK; Hoechstetter, MA; Buchner, M et al.
Blood Adv (2023)
 Direct Cryo-ET observation of platelet deformation induced by SARS-CoV-2 spike protein
Direct Cryo-ET observation of platelet deformation induced by SARS-CoV-2 spike protein
Kuhn, CC; Basnet, N; Bodakuntla, S et al.
Nat Commun (2023)
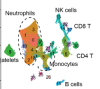 Spleen tyrosine kinase inhibition restores myeloid homeostasis in COVID-19
Spleen tyrosine kinase inhibition restores myeloid homeostasis in COVID-19
Wigerblad, G; Warner, SA; Ramos-Benitez, MJ et al.
Sci Adv (2023)
Serological responses to human virome define clinical outcomes of Italian patients infected with SARS-CoV-2
Wang, L; Candia, J; Ma, L et al.
Int J Biol Sci (2022)
Acute worsening of CADASIL in a patient with COVID-19 infection: illustrative case
Rosenblum, JS; Tunacao, JM; Nazari, MA et al.
J Neurosurg Case Lessons (2022)
 Temporal Improvements in COVID-19 Outcomes for Hospitalized Adults: A Post Hoc Observational Study of Remdesivir Group Participants in the Adaptive COVID-19 Treatment Trial
Temporal Improvements in COVID-19 Outcomes for Hospitalized Adults: A Post Hoc Observational Study of Remdesivir Group Participants in the Adaptive COVID-19 Treatment Trial
Potter, GE; Bonnett, T; Rubenstein, K et al.
Ann Intern Med (2022)
 Intranasal delivery of a rationally attenuated SARS-CoV-2 is immunogenic and protective in Syrian hamsters
Intranasal delivery of a rationally attenuated SARS-CoV-2 is immunogenic and protective in Syrian hamsters
Liu, S; Stauft, CB; Selvaraj, P et al.
Nat Commun (2022)
 Limited cross-variant immune response from SARS-CoV-2 Omicron BA.2 in naïve but not previously infected outpatients
Limited cross-variant immune response from SARS-CoV-2 Omicron BA.2 in naïve but not previously infected outpatients
Lee, HK; Knabl, L; Walter, M et al.
iScience (2022)
 Comprehensive analysis of immune responses in CLL patients after heterologous COVID-19 vaccination
Comprehensive analysis of immune responses in CLL patients after heterologous COVID-19 vaccination
Lee, HK; Hoechstetter, MA; Buchner, M et al.
medRxiv (2022)
 Within-host genetic diversity of SARS-CoV-2 in the context of large-scale hospital-associated genomic surveillance
Within-host genetic diversity of SARS-CoV-2 in the context of large-scale hospital-associated genomic surveillance
Mushegian, A; Long, SW; Olsen, RJ et al.
medRxiv (2022)
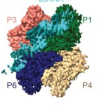 Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease
Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease
Frazier, MN; Wilson, IM; Krahn, JM et al.
Nucleic Acids Res (2022)
 Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries
Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries
Banday, AR; Stanifer, ML; Florez-Vargas, O et al.
Nat Genet (2022)
Clonal Hematopoiesis Is Not Significantly Associated with Covid-19 Disease Severity
Zhou, Y; Shalhoub, RN; Rogers, SN et al.
Blood (2022)
 A Path-Based Analysis of Infected Cell Line and COVID-19 Patient Transcriptome Reveals Novel Potential Targets and Drugs Against SARS-CoV-2
A Path-Based Analysis of Infected Cell Line and COVID-19 Patient Transcriptome Reveals Novel Potential Targets and Drugs Against SARS-CoV-2
Agrawal, P; Sambaturu, N; Olgun, G; Hannenhalli, S; ,
Front Immunol (2022)
 Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants
Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants
Hong, J; Kwon, HJ; Cachau, R et al.
Proc Natl Acad Sci U S A (2022)
 Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein
Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein
Zhao, H; Nguyen, A; Wu, D et al.
PNAS Nexus (2022)
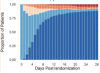 Unraveling the Treatment Effect of Baricitinib on Clinical Progression and Resource Utilization in Hospitalized COVID-19 Patients: Secondary Analysis of the Adaptive COVID-19 Treatment Randomized Trial-2
Unraveling the Treatment Effect of Baricitinib on Clinical Progression and Resource Utilization in Hospitalized COVID-19 Patients: Secondary Analysis of the Adaptive COVID-19 Treatment Randomized Trial-2
Fintzi, J; Bonnett, T; Tebas, P et al.
Open Forum Infect Dis (2022)
 Prior Vaccination Exceeds Prior Infection in Eliciting Innate and Humoral Immune Responses in Omicron Infected Outpatients
Prior Vaccination Exceeds Prior Infection in Eliciting Innate and Humoral Immune Responses in Omicron Infected Outpatients
Lee, HK; Knabl, L; Walter, M et al.
Front Immunol (2022)
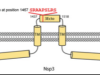 Putative Host-Derived Insertions in the Genomes of Circulating SARS-CoV-2 Variants
Putative Host-Derived Insertions in the Genomes of Circulating SARS-CoV-2 Variants
Yang, Y; Dufault-Thompson, K; Salgado Fontenele, R; Jiang, X; ,
mSystems (2022)
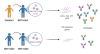 Heterologous ChAdOx1-BNT162b2 vaccination in Korean cohort induces robust immune and antibody responses that includes Omicron
Heterologous ChAdOx1-BNT162b2 vaccination in Korean cohort induces robust immune and antibody responses that includes Omicron
Lee, HK; Go, J; Sung, H et al.
iScience (2022)
 Limited cross-variant immune response from SARS-CoV-2 Omicron BA.2 in naïve but not previously infected outpatients
Limited cross-variant immune response from SARS-CoV-2 Omicron BA.2 in naïve but not previously infected outpatients
Lee, HK; Knabl, L; Walter, M et al.
medRxiv (2022)
 Camel nanobodies broadly neutralize SARS-CoV-2 variants
Camel nanobodies broadly neutralize SARS-CoV-2 variants
Hong, J; Kwon, HJ; Cachau, R et al.
bioRxiv (2021)
 SARS-CoV-2 introductions and early dynamics of the epidemic in Portugal
SARS-CoV-2 introductions and early dynamics of the epidemic in Portugal
Borges, V; Isidro, J; Trovão, NS et al.
Commun Med (Lond) (2022)
 Synthetic lethality-based prediction of anti-SARS-CoV-2 targets
Synthetic lethality-based prediction of anti-SARS-CoV-2 targets
Pal, LR; Cheng, K; Nair, NU et al.
iScience (2022)
 BNT162b2 vaccination enhances interferon-JAK-STAT-regulated antiviral programs in COVID-19 patients infected with the SARS-CoV-2 Beta variant
BNT162b2 vaccination enhances interferon-JAK-STAT-regulated antiviral programs in COVID-19 patients infected with the SARS-CoV-2 Beta variant
Knabl, L; Lee, HK; Wieser, M et al.
Commun Med (Lond) (2022)
 Evolutionary history and introduction of SARS-CoV-2 Alpha VOC/B.1.1.7 in Pakistan through international travelers
Evolutionary history and introduction of SARS-CoV-2 Alpha VOC/B.1.1.7 in Pakistan through international travelers
Nasir, A; Bukhari, AR; Trovão, NS et al.
Virus Evol (2022)
 mRNA vaccination in octogenarians 15 and 20 months after recovery from COVID-19 elicits robust immune and antibody responses that include Omicron
mRNA vaccination in octogenarians 15 and 20 months after recovery from COVID-19 elicits robust immune and antibody responses that include Omicron
Lee, HK; Knabl, L; Moliva, JI et al.
Cell Rep (2022)
 Yearlong COVID-19 Infection Reveals Within-Host Evolution of SARS-CoV-2 in a Patient With B-Cell Depletion
Yearlong COVID-19 Infection Reveals Within-Host Evolution of SARS-CoV-2 in a Patient With B-Cell Depletion
Nussenblatt, V; Roder, AE; Das, S et al.
J Infect Dis (2022)
 Parsing Immune Correlates of Protection Against SARS-CoV-2 from Biomedical Literature
Parsing Immune Correlates of Protection Against SARS-CoV-2 from Biomedical Literature
Foote, SL; Jones, S; Lockmuller, J et al.
AMIA Annu Symp Proc (2021)
 Preintubation Sequential Organ Failure Assessment Score for Predicting COVID-19 Mortality: External Validation Using Electronic Health Record From 86 U.S. Healthcare Systems to Appraise Current Ventilator Triage Algorithms
Preintubation Sequential Organ Failure Assessment Score for Predicting COVID-19 Mortality: External Validation Using Electronic Health Record From 86 U.S. Healthcare Systems to Appraise Current Ventilator Triage Algorithms
Keller, MB; Wang, J; Nason, M et al.
Crit Care Med (2022)
 A year of COVID-19 GWAS results from the GRASP portal reveals potential genetic risk factors
A year of COVID-19 GWAS results from the GRASP portal reveals potential genetic risk factors
Thibord, F; Chan, MV; Chen, MH; Johnson, AD; ,
HGG Adv (2022)
 Immune transcriptome analysis of COVID-19 patients infected with SARS-CoV-2 variants carrying the E484K escape mutation identifies a distinct gene module
Immune transcriptome analysis of COVID-19 patients infected with SARS-CoV-2 variants carrying the E484K escape mutation identifies a distinct gene module
Lee, HK; Knabl, L; Knabl, L et al.
Sci Rep (2022)
 SARS-CoV-2 infection in free-ranging white-tailed deer
SARS-CoV-2 infection in free-ranging white-tailed deer
Hale, VL; Dennis, PM; McBride, DS et al.
Nature (2022)
 Epidemiology and genetic diversity of SARS-CoV-2 lineages circulating in Africa
Epidemiology and genetic diversity of SARS-CoV-2 lineages circulating in Africa
Okoh, OS; Nii-Trebi, NI; Jakkari, A et al.
iScience (2022)
 Putative host-derived insertions in the genome of circulating SARS-CoV-2 variants
Putative host-derived insertions in the genome of circulating SARS-CoV-2 variants
Yang, Y; Dufault-Thompson, K; Fontenele, RS; Jiang, X; ,
bioRxiv (2022)
 Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern
Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern
Li, W; Chen, Y; Prévost, J et al.
Cell Rep (2022)
 Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U
Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U
Frazier, MN; Dillard, LB; Krahn, JM et al.
Nucleic Acids Res (2021)
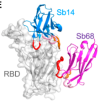 Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction
Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction
Ahmad, J; Jiang, J; Boyd, LF et al.
J Biol Chem (2021)
 Genome-scale metabolic modeling reveals SARS-CoV-2-induced metabolic changes and antiviral targets
Genome-scale metabolic modeling reveals SARS-CoV-2-induced metabolic changes and antiviral targets
Cheng, K; Martin-Sancho, L; Pal, LR et al.
Mol Syst Biol (2021)
 Year-long COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B cell depletion
Year-long COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B cell depletion
Nussenblatt, V; Roder, AE; Das, S et al.
medRxiv (2021)
 Deconstructing the Treatment Effect of Remdesivir in the Adaptive COVID-19 Treatment Trial-1: Implications for Critical Care Resource Utilization
Deconstructing the Treatment Effect of Remdesivir in the Adaptive COVID-19 Treatment Trial-1: Implications for Critical Care Resource Utilization
Fintzi, J; Bonnett, T; Sweeney, DA et al.
Clin Infect Dis (2021)
Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome
Sulaiman, I; Chung, M; Angel, L et al.
Nat Microbiol (2021)
The NIH Lipo-COVID Study: A Pilot NMR Investigation of Lipoprotein Subfractions and Other Metabolites in Patients with Severe COVID-19
Ballout, RA; Kong, H; Sampson, M et al.
Biomedicines (2021)
Robust immune response to the BNT162b mRNA vaccine in an elderly population vaccinated 15 months after recovery from COVID-19
Lee, HK; Knabl, L; Knabl, L et al.
medRxiv (2021)
Regulation of the Dimerization and Activity of SARS-CoV-2 Main Protease through Reversible Glutathionylation of Cysteine 300
Davis, DA; Bulut, H; Shrestha, P et al.
mBio (2021)
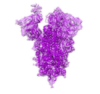 SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity
SARS-CoV-2 S2P spike ages through distinct states with altered immunogenicity
Olia, AS; Tsybovsky, Y; Chen, SJ et al.
J Biol Chem (2021)
 Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants
Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants
Xu, J; Xu, K; Jung, S et al.
Nature (2021)
 Genome-wide covariation in SARS-CoV-2
Genome-wide covariation in SARS-CoV-2
Cresswell-Clay, E; Periwal, V; ,
Math Biosci (2021)
 JAK inhibitors dampen activation of interferon-activated transcriptomes and the SARS-CoV-2 receptor ACE2 in human renal proximal tubules
JAK inhibitors dampen activation of interferon-activated transcriptomes and the SARS-CoV-2 receptor ACE2 in human renal proximal tubules
Jankowski, J; Lee, HK; Wilflingseder, J; Hennighausen, L; ,
iScience (2021)
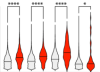 Host-Virus Chimeric Events in SARS-CoV-2-Infected Cells Are Infrequent and Artifactual
Host-Virus Chimeric Events in SARS-CoV-2-Infected Cells Are Infrequent and Artifactual
Yan, B; Chakravorty, S; Mirabelli, C et al.
J Virol (2021)
 Genetic regulation of OAS1 nonsense-mediated decay underlies association with risk of severe COVID-19
Genetic regulation of OAS1 nonsense-mediated decay underlies association with risk of severe COVID-19
Banday, AR; Stanifer, ML; Florez-Vargas, O et al.
medRxiv (2021)
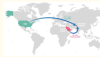 Genetic and evolutionary analysis of SARS-CoV-2 circulating in the region surrounding Islamabad, Pakistan
Genetic and evolutionary analysis of SARS-CoV-2 circulating in the region surrounding Islamabad, Pakistan
Tamim, S; Trovao, NS; Thielen, P et al.
Infect Genet Evol (2021)
 Assessing vaccine durability in randomized trials following placebo crossover
Assessing vaccine durability in randomized trials following placebo crossover
Fintzi, J; Follmann, D; ,
Stat Med (2021)
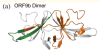 Predictable fold switching by the SARS-CoV-2 protein ORF9b
Predictable fold switching by the SARS-CoV-2 protein ORF9b
Porter, LL
Protein Sci (2021)
 JAK inhibitors dampen activation of interferon-stimulated transcription of ACE2 isoforms in human airway epithelial cells
JAK inhibitors dampen activation of interferon-stimulated transcription of ACE2 isoforms in human airway epithelial cells
Lee, HK; Jung, O; Hennighausen, L; ,
Commun Biol (2021)
 Immune transcriptomes from hospitalized patients infected with the SARS-CoV-2 variants B.1.1.7 and B.1.1.7 carrying the E484K escape mutation
Immune transcriptomes from hospitalized patients infected with the SARS-CoV-2 variants B.1.1.7 and B.1.1.7 carrying the E484K escape mutation
Lee, HK; Knabl, L; Knabl, L et al.
medRxiv (2021)
 Impact of BNT162b first vaccination on the immune transcriptome of elderly patients infected with the B.1.351 SARS-CoV-2 variant
Impact of BNT162b first vaccination on the immune transcriptome of elderly patients infected with the B.1.351 SARS-CoV-2 variant
Knabl, L; Lee, HK; Wieser, M et al.
medRxiv (2021)
 COVID-19-CT-CXR: A Freely Accessible and Weakly Labeled Chest X-Ray and CT Image Collection on COVID-19 From Biomedical Literature
COVID-19-CT-CXR: A Freely Accessible and Weakly Labeled Chest X-Ray and CT Image Collection on COVID-19 From Biomedical Literature
Peng, Y; Tang, Y; Lee, S et al.
IEEE Trans Big Data (2021)
Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury
Andargie, TE; Tsuji, N; Seifuddin, F et al.
JCI Insight (2021)
 Social Heterogeneity Drives Complex Patterns of the COVID-19 Pandemic: Insights From a Novel Stochastic Heterogeneous Epidemic Model (SHEM)
Social Heterogeneity Drives Complex Patterns of the COVID-19 Pandemic: Insights From a Novel Stochastic Heterogeneous Epidemic Model (SHEM)
Alexander V. Maltsev & Michael D. Stern*
Front. Phys. (2021)
 FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist
FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist
Baena, V; Conrad, R; Friday, P et al.
Viruses (2021)
COVID-19 reopening strategies at the county level in the face of uncertainty: Multiple Models for Outbreak Decision Support
Shea, K; Borchering, RK; Probert, WJM et al.
medRxiv (2020)
 SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation
SARS-CoV-2 drives JAK1/2-dependent local complement hyperactivation
Yan, B; Freiwald, T; Chauss, D et al.
Sci Immunol (2021)
 Network Analysis and Transcriptome Profiling Identify Autophagic and Mitochondrial Dysfunctions in SARS-CoV-2 Infection
Network Analysis and Transcriptome Profiling Identify Autophagic and Mitochondrial Dysfunctions in SARS-CoV-2 Infection
Singh, K; Chen, YC; Hassanzadeh, S et al.
Front Genet (2021)
 SARS-CoV-2 infection of the oral cavity and saliva
SARS-CoV-2 infection of the oral cavity and saliva
Huang, N; Pérez, P; Kato, T et al.
Nat Med (2021)
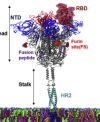 Exploring dynamics and network analysis of spike glycoprotein of SARS-COV-2
Exploring dynamics and network analysis of spike glycoprotein of SARS-COV-2
Ghorbani, M; Brooks, BR; Klauda, JB; ,
Biophys J (2021)
 Genomic diversity of SARS-CoV-2 during early introduction into the Baltimore-Washington metropolitan area
Genomic diversity of SARS-CoV-2 during early introduction into the Baltimore-Washington metropolitan area
Thielen, PM; Wohl, S; Mehoke, T et al.
JCI Insight (2021)
 Immune transcriptomes of highly exposed SARS-CoV-2 asymptomatic seropositive versus seronegative individuals from the Ischgl community
Immune transcriptomes of highly exposed SARS-CoV-2 asymptomatic seropositive versus seronegative individuals from the Ischgl community
Lee, HK; Knabl, L; Pipperger, L et al.
Sci Rep (2021)
 Inborn errors of type I IFN immunity in patients with life-threatening COVID-19
Inborn errors of type I IFN immunity in patients with life-threatening COVID-19
Zhang, Q; Bastard, P; Liu, Z et al.
Science (2020)
Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation
Zhang, L; Mann, M; Syed, Z et al.
bioRxiv (2021)
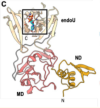
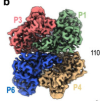 Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics
Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics
Pillon, MC; Frazier, MN; Dillard, LB et al.
Nat Commun (2021)
 Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15
Cryo-EM Structures of the SARS-CoV-2 Endoribonuclease Nsp15
Pillon, MC; Frazier, MN; Dillard, LB et al.
bioRxiv (2020)
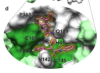 A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication
A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication
Hattori, SI; Higashi-Kuwata, N; Hayashi, H et al.
Nat Commun (2021)
 Interferon-regulated genetic programs and JAK/STAT pathway activate the intronic promoter of the short ACE2 isoform in renal proximal tubules
Interferon-regulated genetic programs and JAK/STAT pathway activate the intronic promoter of the short ACE2 isoform in renal proximal tubules
Jankowski, J; Lee, HK; Wilflingseder, J; Hennighausen, L; ,
bioRxiv (2021)
Integrated Single-Cell Atlases Reveal an Oral SARS-CoV-2 Infection and Transmission Axis
Huang, N; Perez, P; Kato, T et al.
medRxiv (2020)
 Assessing Durability of Vaccine Effect Following Blinded Crossover in COVID-19 Vaccine Efficacy Trials
Assessing Durability of Vaccine Effect Following Blinded Crossover in COVID-19 Vaccine Efficacy Trials
Follmann, D; Fintzi, J; Fay, MP et al.
medRxiv (2020)
Activation of Interferon-Stimulated Transcriptomes and ACE2 Isoforms in Human Airway Epithelium Is Curbed by Janus Kinase Inhibitors
Lee, HK; Jung, O; Hennighausen, L; ,
Res Sq (2020)
 An immune-based biomarker signature is associated with mortality in COVID-19 patients
An immune-based biomarker signature is associated with mortality in COVID-19 patients
Abers, MS; Delmonte, OM; Ricotta, EE et al.
JCI Insight (2020)
 Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination
Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination
Yang, Y; Yan, W; Hall, AB; Jiang, X; ,
Mol Biol Evol (2020)
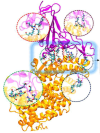 Critical Sequence Hotspots for Binding of Novel Coronavirus to Angiotensin Converter Enzyme as Evaluated by Molecular Simulations
Critical Sequence Hotspots for Binding of Novel Coronavirus to Angiotensin Converter Enzyme as Evaluated by Molecular Simulations
Ghorbani, M; Brooks, BR; Klauda, JB; ,
J Phys Chem B (2020)
 Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor
Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor
Onabajo, OO; Banday, AR; Stanifer, ML et al.
Nat Genet (2020)
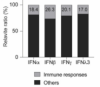 Activation of ACE2 and interferon-stimulated transcriptomes in human airway epithelium is curbed by Janus Kinase inhibitors
Activation of ACE2 and interferon-stimulated transcriptomes in human airway epithelium is curbed by Janus Kinase inhibitors
Lee, HK; Jung, O; Hennighausen, L; ,
bioRxiv (2020)
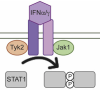 Activation of the SARS-CoV-2 Receptor Ace2 through JAK/STAT-Dependent Enhancers during Pregnancy
Activation of the SARS-CoV-2 Receptor Ace2 through JAK/STAT-Dependent Enhancers during Pregnancy
Hennighausen, L; Lee, HK; ,
Cell Rep (2020)
 Activation of the antiviral factor RNase L triggers translation of non-coding mRNA sequences
Activation of the antiviral factor RNase L triggers translation of non-coding mRNA sequences
Karasik A, Jones D, DePass A, Guydosh N
bioRxiv (2020)
 In silico Drug Repurposing to combat COVID-19 based on Pharmacogenomics of Patient Transcriptomic Data
In silico Drug Repurposing to combat COVID-19 based on Pharmacogenomics of Patient Transcriptomic Data
Das, S; Camphausen, K; Shankavaram, U; ,
Res Sq (2020)
 Using multiple data streams to estimate and forecast SARS-CoV-2 transmission dynamics, with application to the virus spread in Orange County, California
Using multiple data streams to estimate and forecast SARS-CoV-2 transmission dynamics, with application to the virus spread in Orange County, California
Fintzi, J; Bayer, D; Goldstein, I et al.
ArXiv (2020)
 GRL-0920, an Indole Chloropyridinyl Ester, Completely Blocks SARS-CoV-2 Infection
GRL-0920, an Indole Chloropyridinyl Ester, Completely Blocks SARS-CoV-2 Infection
Hattori, SI; Higshi-Kuwata, N; Raghavaiah, J et al.
mBio (2020)
 Genomic Diversity of SARS-CoV-2 During Early Introduction into the United States National Capital Region
Genomic Diversity of SARS-CoV-2 During Early Introduction into the United States National Capital Region
Theilen PM; Wohl S; Mehoke T; et al
medRxiv (2020)
 An autocrine Vitamin D-driven Th1 shutdown program can be exploited for COVID-19
An autocrine Vitamin D-driven Th1 shutdown program can be exploited for COVID-19
McGregor, R; Chauss, D; Freiwald, T et al.
bioRxiv (2020)
 In vitro and in vivo identification of clinically approved drugs that modify ACE2 expression
In vitro and in vivo identification of clinically approved drugs that modify ACE2 expression
Sinha, S; Cheng, K; Schäffer, AA; Aldape, K; Schiff, E; Ruppin, E;
Mol. Syst. Biol. (2020)
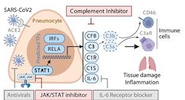 SARS-CoV2 drives JAK1/2-dependent local and systemic complement hyper-activation
SARS-CoV2 drives JAK1/2-dependent local and systemic complement hyper-activation
Yan, B; Freiwald, T; et al
researchsquare (2020)
 Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing
Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing
Riva, L; Yuan, S; Yin, X et al.
Nature (2020)
 Statistical Physics of Epidemic on Network Predictions for SARS-CoV-2 Parameters
Statistical Physics of Epidemic on Network Predictions for SARS-CoV-2 Parameters
Han, J; Cresswell-Clay, EC; Periwal, V
ArXiv (2020)
 Critical Sequence Hot-spots for Binding of nCOV-2019 to ACE2 as Evaluated by Molecular Simulations
Critical Sequence Hot-spots for Binding of nCOV-2019 to ACE2 as Evaluated by Molecular Simulations
Ghorbani, M; Brooks, BR; Klauda JB
bioRxiv (2020)
 Global prediction of unreported SARS-CoV2 infection from observed COVID-19 cases
Global prediction of unreported SARS-CoV2 infection from observed COVID-19 cases
Chow, CC; Chang, JC; Gerkin, RC; Vattikuti, S;
medRxiv (2020)
 Activation of the SARS-CoV-2 receptor Ace2 by cytokines through pan JAK-STAT enhancers
Activation of the SARS-CoV-2 receptor Ace2 by cytokines through pan JAK-STAT enhancers
Hennighausen, L; Lee, HK;
bioRxiv (2020)
Preprints are complete and public drafts of scientific documents, not yet peer reviewed. (More about preprints)
0 preprint papers




