The NIH HPC group plans, manages and supports
high-performance computing systems specifically for use by the
intramural NIH community. These systems include
Biowulf,
a 105,000+ processor
Linux cluster;
Helix, an interactive system for
file transfer and management, and
Helixweb, which provides a number of web-based
scientific tools. We provide access to a wide range of computational
applications for genomics, molecular and structural biology, mathematical and
graphical analysis, image analysis, and other scientific fields.
The continued growth and support of NIH's Biowulf cluster is dependent upon
its demonstrable value to the NIH Intramural Research Program. If you publish
research that involved significant use of Biowulf, please cite the cluster.
Suggested citation text:
This work utilized the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
|
Quick Links
Biowulf Utilization
Thursday, February 26th, 2026
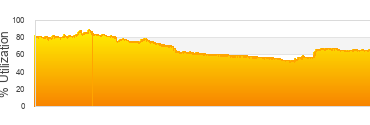
|
Last 24 hrs
|
|
112,406 jobs submitted
85,388 jobs completed
2,708,239 CPU hrs used
|
25 NIH Institutes
339 Principal Investigators
726 users
|
Announcements
|
Recent Papers that used Biowulf & HPC Resources
|
Molecular basis for the regulation of membrane proteins through preferential lipid solvation
Bernhardt, N; Ozturk, TN; Zhang, S et al.
Nat Chem Biol
, DOI://10.1038/s41589-025-02032-w (2025)
The Time-Dependent Association Between Irritable Bowel Syndrome and All-Cause and Cause-Specific Mortality in the NIH-AARP Cohort Study
Gutiérrez-Torres, DS; Li, F; Liao, LM et al.
Am J Gastroenterol
, DOI://10.14309/ajg.0000000000003861 (2025)
Taxonomic-Level Protein Quantification in Metaproteomics Using a Biomass-Constrained Expectation-Maximization Approach
Alves, G; Hamaneh, MB; Ogurtsov, AY; Yu, YK; ,
J Am Soc Mass Spectrom
, DOI://10.1021/jasms.5c00332 (2026)
Spatial Analysis of Hereditary Diffuse Gastric Cancer Reveals Indolent Phenotype of Signet Ring Cell Precursors
Gallanis, AF; Gamble, LA; Oguz, C et al.
Mol Cancer Res
, DOI://10.1158/1541-7786.MCR-24-1039 (2025)
An insight into the ovary and midgut transcriptome of Dermacentor nitens tick
Lu, S; Bosio, CF; Andrade-Silva, V et al.
Ticks Tick Borne Dis
, DOI://10.1016/j.ttbdis.2025.102600 (2026)
Children's state anxiety before MRI scanning and resting state functional connectivity in large scale brain networks
Qamar, P; Díaz, DE; Benson, BE et al.
Sci Rep
, DOI://10.1038/s41598-025-34410-8 (2026)
PCLIPtools: a robust framework for identifying RNA-protein interaction sites from PAR-CLIP experiments
Polash, AH; Hafner, M; ,
Nucleic Acids Res
, DOI://10.1093/nar/gkag062 (2026)
Eos plays a critical role in Treg homeostasis and modulates the function of recirculating thymic Tregs in the control of Treg development
Xie, X; Thornton, AM; Tanwar, S et al.
Cell Rep
, DOI://10.1016/j.celrep.2025.116838 (2026)
|
|
 All Services Operational
All Services Operational All Services Operational
All Services Operational