Chapter 9 Calculate genotype posteriors
9.1 Brief introduction
After we filtered our callset, we use extra information like pedigree and allele frequencies in relevant populations to refine the genotype assignments.
9.2 Benchmarks
We did benchmarks on the performance of CalculateGenotypePosteriors with
different numbers of CPUs and memory. As show in figure 9.1,
the runtime was not reduced with increasing threads.
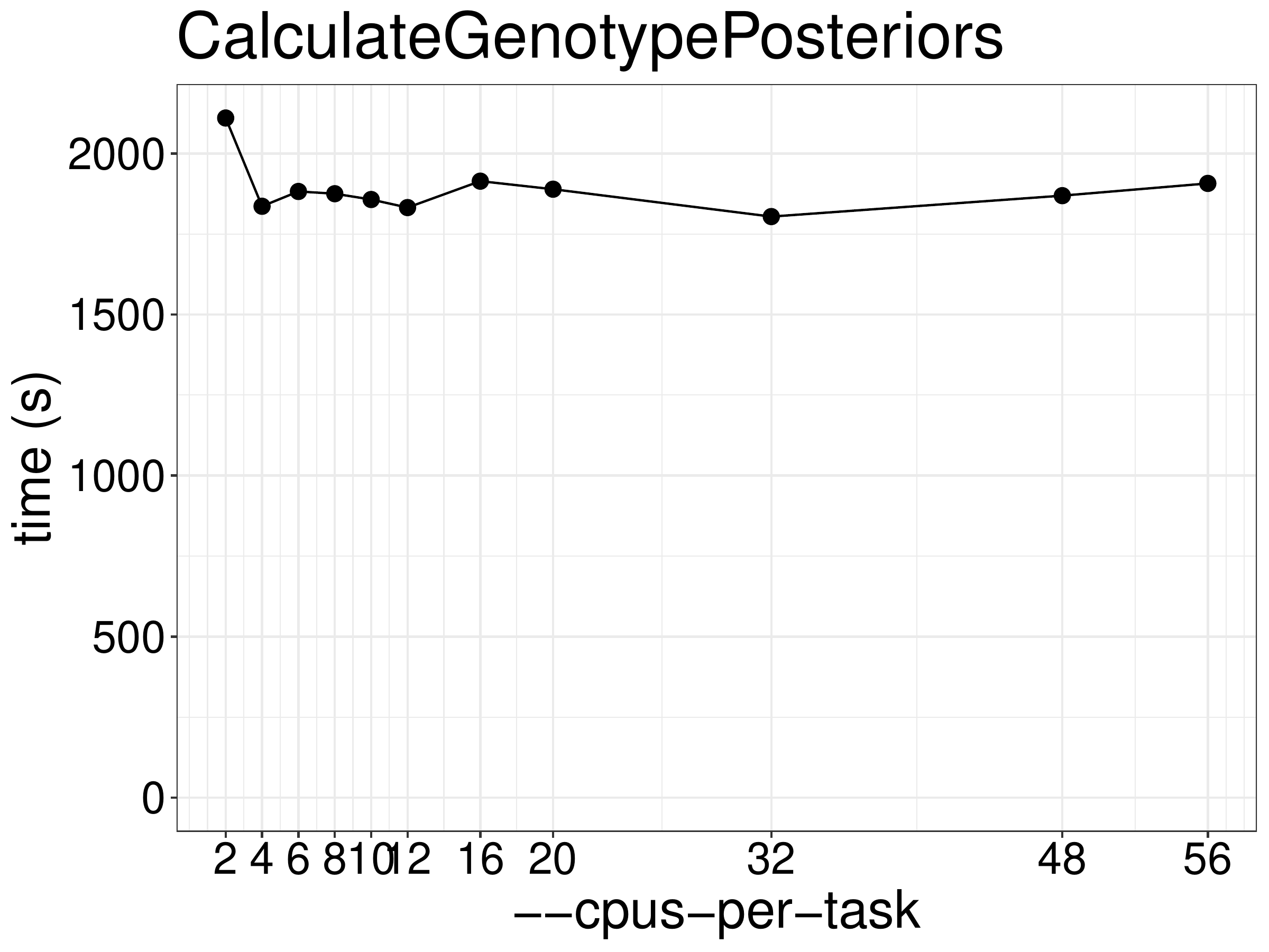
Figure 9.1: Runtime of CalculateGenotypePosteriors as a function of the number of threads
We normally recommend running jobs with 70%-80% efficiency. Based on the efficiency calculated from the benchmarks above (figure 9.2)
we recommend not running CalculateGenotypePosteriors with more than 2 threads.
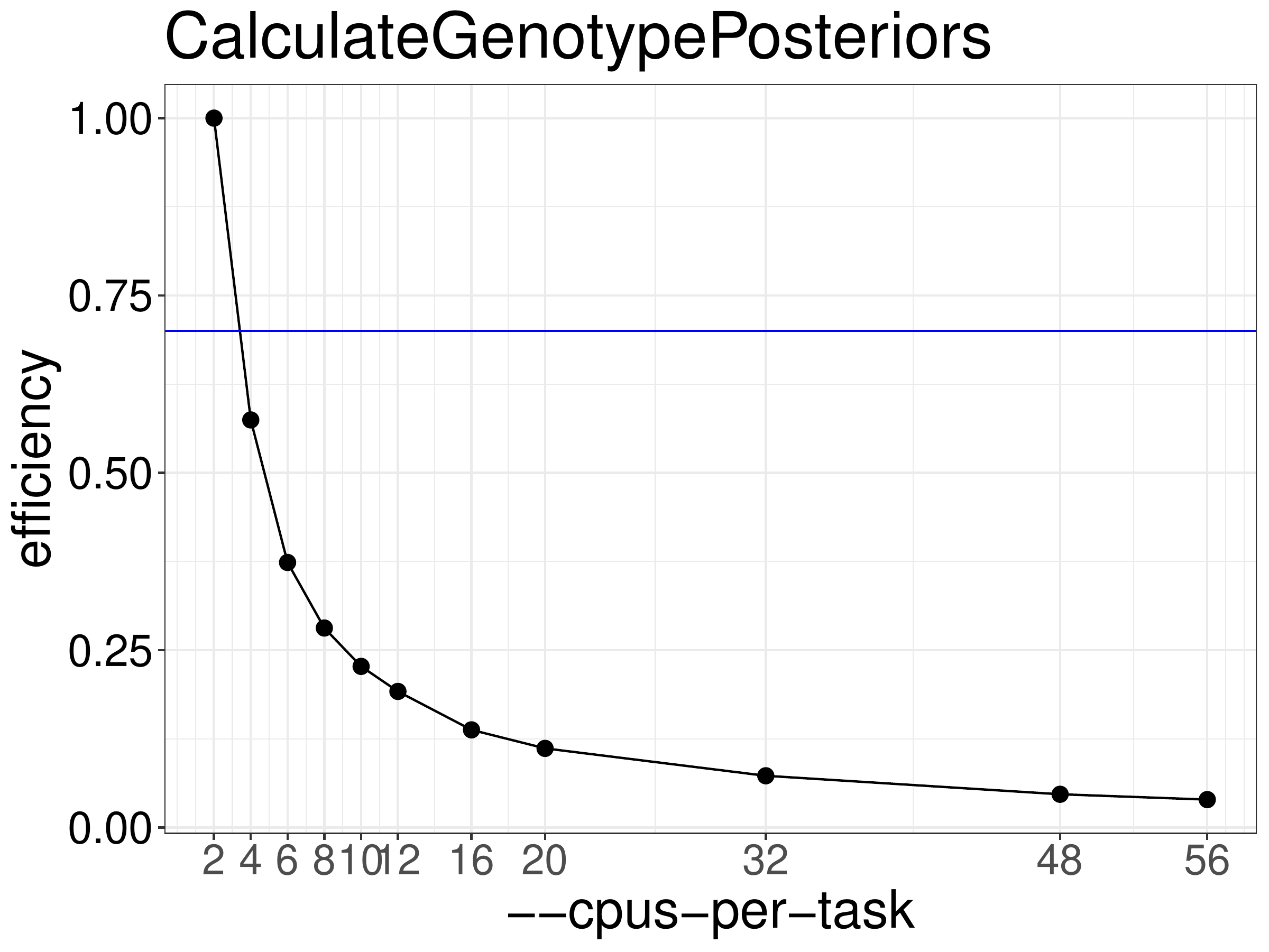
Figure 9.2: Efficiency of CalculateGenotypePosteriors as a function of the number of threads
Increasing memory didn’t improve the performance (figure 9.3).
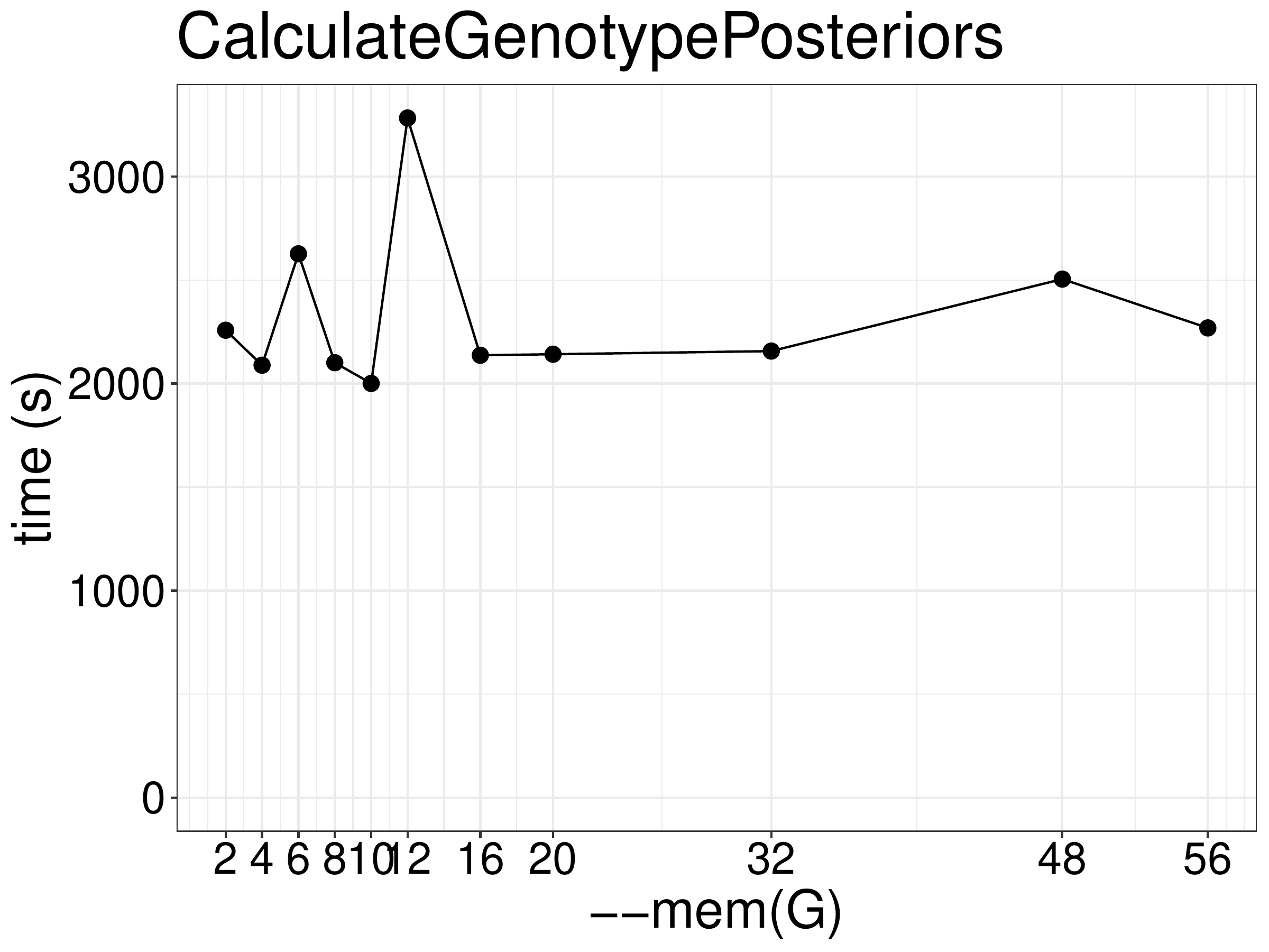
Figure 9.3: Runtime of CalculateGenotypePosteriors as a function of the number of threads
9.3 Optimized script
#! /bin/bash
set -euo
module load GATK/4.3.0.0
cd data/;
gatk --java-options "-Djava.io.tmpdir=/lscratch/$SLURM_JOBID -Xms2G -Xmx2G -XX:ParallelGCThreads=2" \
CalculateGenotypePosteriors \
-V indel.SNP.recalibrated_99.9.vcf.gz \
-ped trio_pedigree.ped \
--supporting-callsets af-only-gnomad.hg38.vcf.gz \
-O trio_refined_99.9.vcf.gzJob submission:
sbatch --cpus-per-task=2 --mem=2G --gres=lscratch:100 --time=2:00:00 09-GATK_CalculateGenotypePosteriors_99.9.shNote:
- There are multiple filters could be applied, it depends on your research needs.