MONAI Label is a free and open-source platform that facilitates the development of AI-based applications that aim at reducing the time required to annotate 3D medical image datasets. It allows researchers to readily deploy their apps as services, which can be made available to clinicians via their preferred user-interface. Currently, MONAI Label readily supports locally installed (3DSlicer) and web-based (OHIF) frontends, and offers two Active learning strategies to facilitate and speed up the training of segmentation algorithms. MONAI Label allows researchers to make incremental improvements to their labeling apps by making them available to other researchers and clinicians alike.
Interactive use of MONAILabel requires a
graphical X11 connection.
Both NX and MobaXterm work well for Windows users,
while XQuartz works well for Mac users.
[user@biowulf]$ sinteractive --mem=20g --gres=gpu:k80:1,lscratch:10 -c4 [user@cn0861 ~]$ module load monailabel [+] Loading singularity 3.8.5-1 on cn4206 [+] Loading cuDNN/7.6.5/CUDA-10.2 libraries... [+] Loading CUDA Toolkit 10.2.89 ... [+] Loading monailabel 0.4.0rc4
[user@cn0861 ~]$ monailabel -h
...
monailabel/0.4.0rc4/lib/python3.9/site-packages:/usr/local/Anaconda/envs/py3.9/lib/python3.9/site-packages
usage: monailabel [-h] {start_server,apps,datasets,plugins} ...
positional arguments:
{start_server,apps,datasets,plugins}
sub-command help
start_server Start Application Server
apps list or download sample apps
datasets list or download sample datasets
plugins list or download viewer plugins
optional arguments:
-h, --help show this help message and exit
[user@cn0861 ~]$ PEGASAS pathway -h
usage: PEGASAS pathway [-h] [-o OUT_DIR] [-n NUM_INTERVAL] [--plotting]
geneExpbySample geneSignatureList groupInfo
required arguments:
geneExpbySample A TSV format matrix of gene expression values (FPKM,
TPM, etc.) where each row is one sample and each
column is one gene.
geneSignatureList One or multiple gene signature sets of pathway of
interest in the format of 'gmt' (see MSigDB webset).
groupInfo A TSV format file provides patient ID and
phenotype/disease stage in each row.
optional arguments:
-h, --help show this help message and exit
-o OUT_DIR, --out-dir OUT_DIR
Output folder name of the analysis.
-n NUM_INTERVAL, --num-interval NUM_INTERVAL
Number of KS enrichment calculation processes one
time.
--plotting Making plots to inspect K-S enrichment scores.
[user@cn0861 ~]$ monailabel datasets --download --name Task09_Spleen --output datasets
Task09_Spleen.tar: 1.50GB [00:49, 32.6MB/s]
2022-06-01 07:56:00,592 - INFO - Downloaded: datasets/Task09_Spleen.tar
2022-06-01 07:56:04,484 - INFO - Verified 'Task09_Spleen.tar', md5: 410d4a301da4e5b2f6f86ec3ddba524e.
2022-06-01 07:56:04,485 - INFO - Writing into directory: /gpfs/gsfs7/users/user/MONAILabel/datasets.
Task09_Spleen is downloaded at: datasets/Task09_Spleen
[user@cn0861 ~]$ monailabel apps --download --name radiology --output apps
radiology is copied at: /gpfs/gsfs7/users/user/MONAILabel/apps/radiology
[user@cn0861 ~]$ firefox &
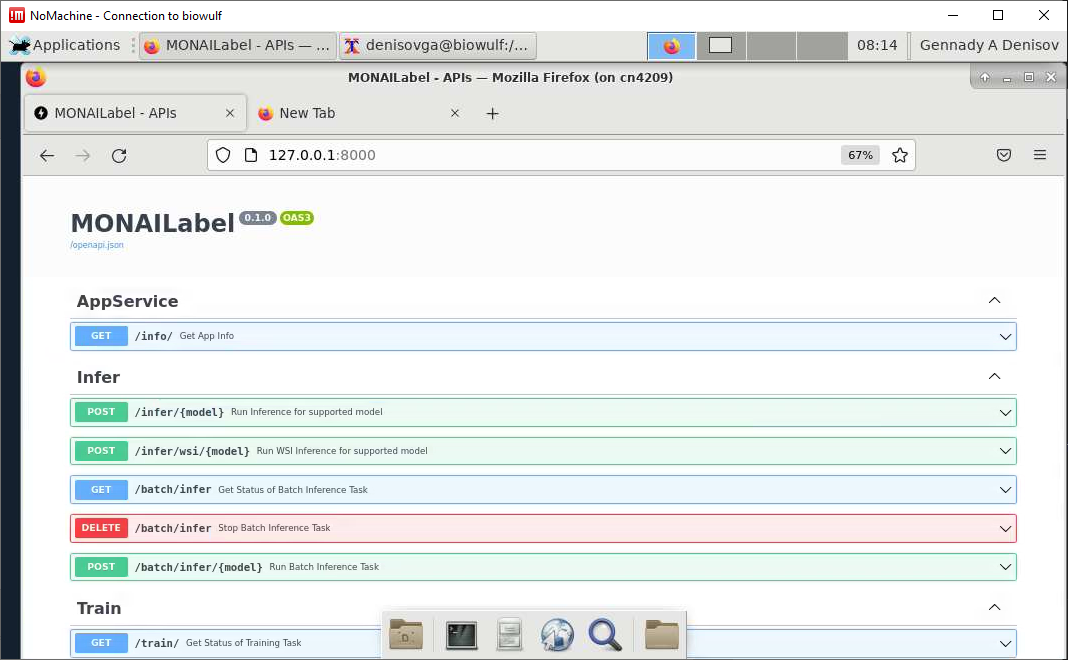
[user@cn0861 ~]$ monailabel start_server --app apps/radiology --studies datasets/Task09_Spleen/imagesTr --conf models deepedit
Navigate the Firefox to the URL: http://127.0.0.1:8000/
[user@cn0861 ~]$ exit salloc.exe: Relinquishing job allocation 46116226