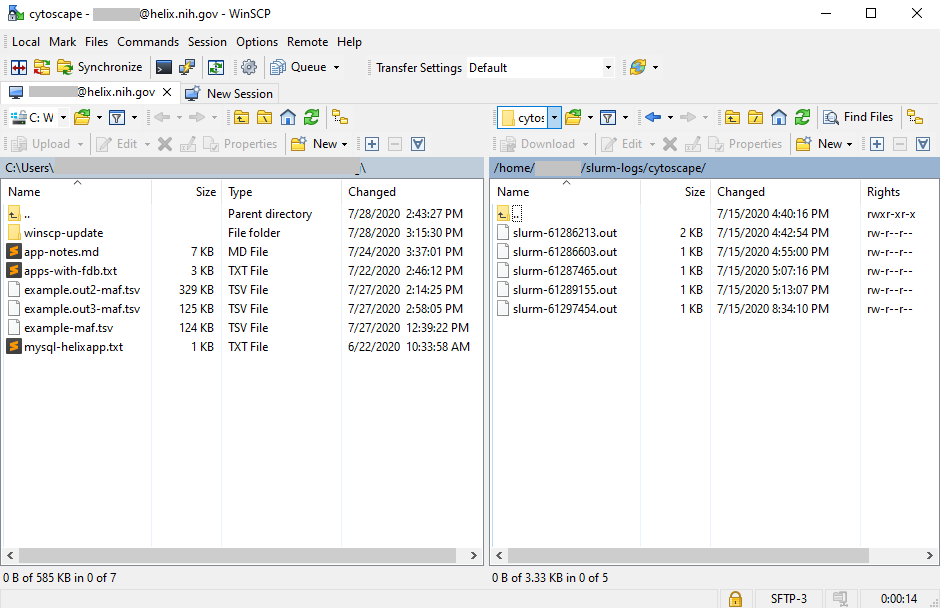
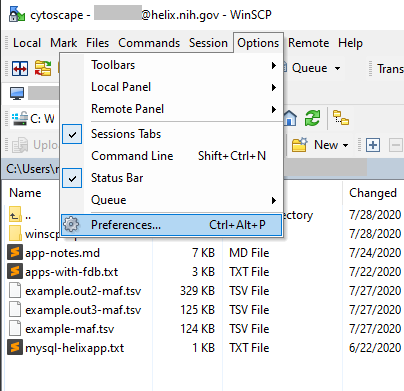
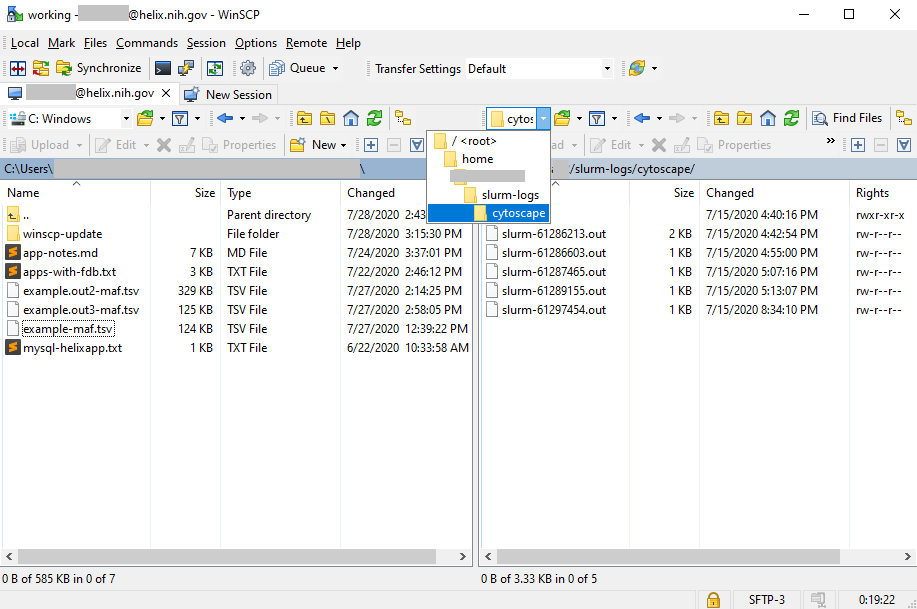
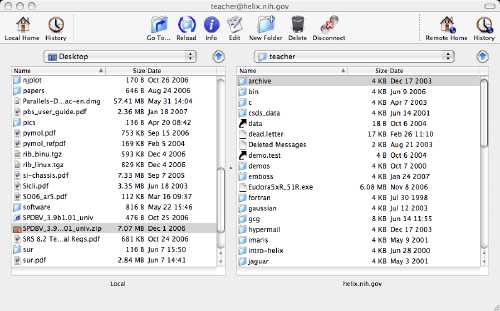
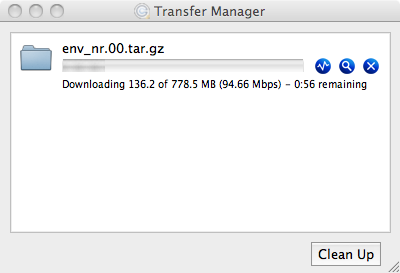
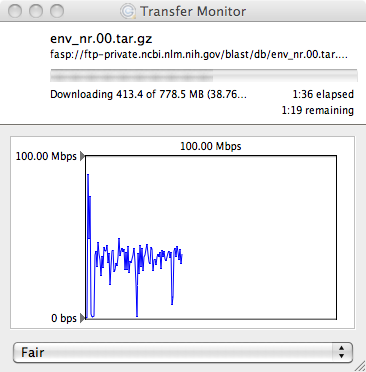
Both psftp and pscp are run through the Windows console (Command Prompt in
start menu), and require the directory to the PuTTY executables be included
in the Path environment variable. This can be done transiently through the console:
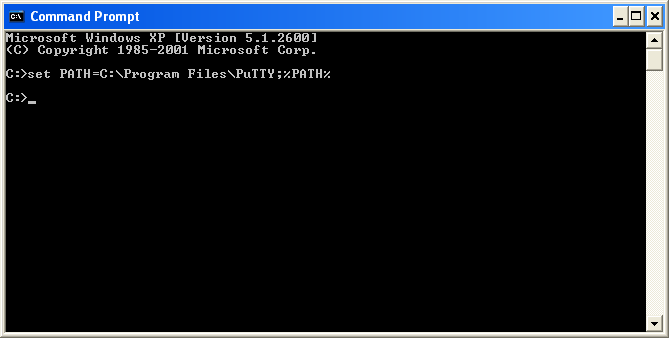
or permanently through the System Control Panel (see here for more information).
pscp
Secure Copy (pscp) is a command line mechanism for copying files to and from remote systems.
From the console, type 'pscp'. This will bring up a help menu showing all the options for pscp.
PuTTY Secure Copy client
Release 0.58
Usage: pscp [options] [user@]host:source target
pscp [options] source [source...] [user@]host:target
pscp [options] -ls [user@]host:filespec
Options:
-V print version information and exit
-pgpfp print PGP key fingerprints and exit
-p preserve file attributes
-q quiet, don't show statistics
-r copy directories recursively
-v show verbose messages
-load sessname Load settings from saved session
-P port connect to specified port
-l user connect with specified user
-pw passw login with specified password
-1 -2 force use of particular SSH protocol version
-4 -6 force use of IPv4 or IPv6
-C enable compression
-i key private key file for authentication
-batch disable all interactive prompts
-unsafe allow server-side wildcards (DANGEROUS)
-sftp force use of SFTP protocol
-scp force use of SCP protocol
To copy a file from the local Windows machine to a user's home directory on Helix, type
C:> pscp localfile user@helix.nih.gov:/home/user/localfile
You will be prompted for your NIH login password, then the file will be copied.
To do the reverse, i.e. copy a remote file from helix to the local Windows machine, type
C:> pscp user@helix.nih.gov:/home/user/remotefile .
(you must include a '.' to retain the same filename, or explicitly give a name
for the remotefile copy).
psftp
Secure FTP (psftp) allows for interactive file transfers between machines in
the same way as good old FTP (non-secure) did.
From the console, type 'psftp'. This will start a sFTP session, but it will
complain that no connection has been made. To transfer a local file to helix, at the psftp prompt type:
psftp> open user@helix.nih.gov
You will again be prompted for a password.
Once a session to helix has been established, the standard FTP commands can be used.
For even more information, see https://www.chiark.greenend.org.uk/~sgtatham/putty/