cisTEM is user-friendly software to process cryo-EM images of macromolecular complexes and obtain high-resolution 3D reconstructions from them.
cisTEM is a beta release. The program may crash for unknown reasons. Please submit issues to the cisTEM Forum.
This application requires a graphical connection. Please use a Graphical Session on HPC OnDemand
There are a few very big problems with running cisTEM on the Biowulf cluster!!
The NIH staff has made available a special QOS for allocating an interactive session for longer than normal. From within the graphical session, allocate a minimal allocation for 5 days:
[user@biowulf]$ sinteractive --qos=cistem --time=5-00:00:00 ... [user@cn3144 ~]$
An interactive session will need to be created within an OnDemand graphical session.
Access to the cistem QOS is only by request. Please contact staff@hpc.nih.gov to be granted access.
Start a cisTEM job on the batch system with appropriate allocations:
[user@cn3144 ~]$ sbatch -p multinode -t 1-00:00 -n 32 --mem-per-cpu=2g -J cisTEM --wrap="ml cisTEM; start_cisTEM" 15465689
NOTES:
Once the batch job begins running, the CisTEM GUI will pop up:

Click on 'Create a new project', fill in the boxes, then click 'Settings', then 'Import'. There should be a file whose name matches the jobid (noted above). This is the run profile text file:
[user@cn3144 ~]$ ls *.txt 15465689_run_profile.txt
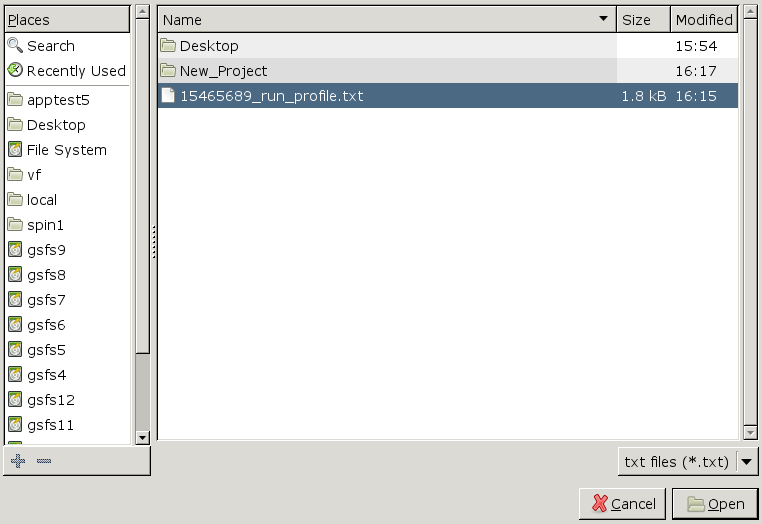
Click 'Open'. Now you have a custom run profile specifically for the current batch job:
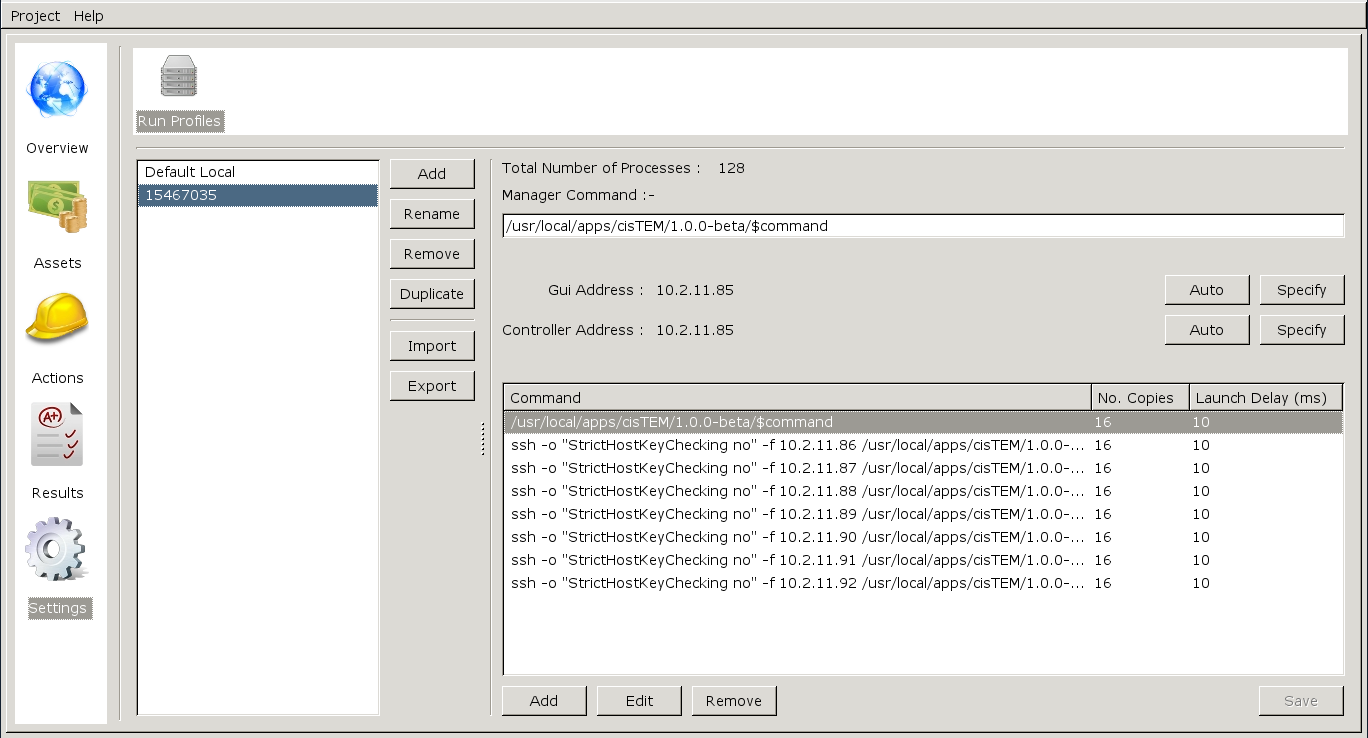
Any tasks run within CisTEM using this run profile will launch commands across the cpus allocated in the batch job.