 [user@cn3144 ~]$ exit
salloc.exe: Relinquishing job allocation 46116226
[user@biowulf ~]$
[user@cn3144 ~]$ exit
salloc.exe: Relinquishing job allocation 46116226
[user@biowulf ~]$
FreeSurfer is a set of automated tools for reconstruction of the brain's cortical surface from structural MRI data, and overlay of functional MRI data onto the reconstructed surface. It was developed at the Martinos Center for Biological Imaging at Harvard.
module load freesurfer ; source $FREESURFER_HOME/SetUpFreeSurfer.sh
Allocate an interactive session with 5 GB of local disk and run the program. Sample session below::
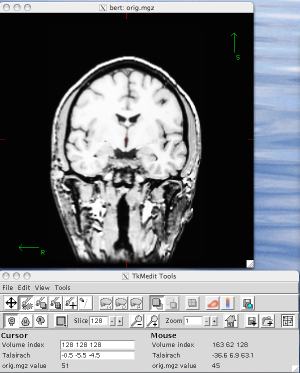
[user@biowulf]$ sinteractive --gres=lscratch:5 salloc.exe: Pending job allocation 46116226 salloc.exe: job 46116226 queued and waiting for resources salloc.exe: job 46116226 has been allocated resources salloc.exe: Granted job allocation 46116226 salloc.exe: Waiting for resource configuration salloc.exe: Nodes cn3144 are ready for job [user@cn3144 ~]$ module load freesurfer [+] Loading Freesurfer 6.0.0 ... [+] Bash users should now type: source $FREESURFER_HOME/SetUpFreeSurfer.sh [+] Csh users should now type: source $FREESURFER_HOME/SetUpFreeSurfer.csh [user@cn3144 ~]$ source $FREESURFER_HOME/SetUpFreeSurfer.sh -------- freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.0-2beb96c -------- Setting up environment for FreeSurfer/FS-FAST (and FSL) FREESURFER_HOME /usr/local/apps/freesurfer/6.0.0 FSFAST_HOME /usr/local/apps/freesurfer/6.0.0/fsfast FSF_OUTPUT_FORMAT nii.gz SUBJECTS_DIR /usr/local/apps/freesurfer/6.0.0/subjects MNI_DIR /usr/local/apps/freesurfer/6.0.0/mni [user@cn3144 ~]$ export tmpdir=/lscratch/$SLURM_JOBID [user@cn3144 ~]$ tkmedit bert orig.mgz Setting subject to bert Reading 0 control points... Reading 0 control points... Reading /usr/local/freesurfer/lib/tcl/tkm_common.tcl Reading /usr/local/freesurfer/lib/tcl/tkm_wrappers.tcl Reading /usr/local/freesurfer/lib/tcl/fsgdfPlot.tcl Reading /usr/local/freesurfer/lib/tcl/tkUtils.tcl[user@cn3144 ~]$ exit salloc.exe: Relinquishing job allocation 46116226 [user@biowulf ~]$
Create a batch input file (e.g. freesurfer.sh). For example:
#!/bin/bash set -e module load freesurfer source $FREESURFER_HOME//SetUpFreeSurfer.sh # set the environment variable tmpdir to local scratch for better performance export tmpdir=/lscratch/$SLURM_JOBID cd /data/user/mydir recon-all -subject hv1 -all
Submit this job using the Slurm sbatch command.
sbatch [--mem=#] --gres=lscratch:5 freesurfer.sh
For swarm jobs, it might be simplest to have the line
module load freesurfer/5.3.0 > /dev/null 2>&1 ; source $FREESURFER_HOME/SetUpFreeSurfer.shin your .bashrc file. Alternatively, you can add this line to each line in your swarm command file.
Create a swarmfile (e.g. freesurfer.swarm). For example:
export tmpdir=/lscratch/$SLURM_JOBID; cd /data/$USER/mydir/; recon-all -subject hv1 -all export tmpdir=/lscratch/$SLURM_JOBID; cd /data/$USER/mydir/; recon-all -subject hv2 -all export tmpdir=/lscratch/$SLURM_JOBID; cd /data/$USER/mydir/; recon-all -subject hv3 -all
Submit this job using the swarm command.
swarm -f freesurfer.swarm [-t #] --gres=lscratch:1where
| -g # | Number of Gigabytes of memory required for each process (1 line in the swarm command file) |
| --gres=lscratch:1 | allocate 1 GB of local disk for each swarm subjob |