gmap-gsnap on Biowulf
Description
GMAP is a tools for rapidly and accurately mapping and aligning cDNA
sequences to genomic sequences.
GSNAP is designed to align short reads from NGS data and allow detection of
short and long range splicing de novo or with a database of know juctions. In
addtion, SNP-tolerant alignements and alignments of reads obtained from
bisulfite treated DNA are implemented.
There may be multiple versions of gmap-gsnap available. An easy way of selecting the
version is to use modules. To see the modules
available, type
module avail gmap-gsnap
To select a module use
module load gmap-gsnap/[version]
where [version] is the version of choice.
gmap-gsnap is a multithreaded application. Make sure to match the
number of cpus requested with the number of threads.
Environment variables set
References
- Thomas D. Wu and Colin K. Watanabe.
GMAP: a genomic mapping and alignment program for mRNA and EST sequences.
Bioinformatics 2005, 21:1859-1875.
Pubmed |
PMC] |
Journal
- Thomas D. Wu and Serban Nacu.
Fast and SNP-tolerant detection of complex variants and splicing in short reads.
Bioinformatics 2010, 26:873-881.
Pubmed |
PMC |
Journal
Documentation
Index files
gmap/gsnap indices are stored under
/fdb/gmap_post20210308/[organism]/[source]/[build]/Sequence/gmap-gsnap
[organism] is the specific organism of interest
(Gallus_gallus, Rattus_norvegicus, etc.)[source] is the source for the sequence (NCBI,
Ensembl, UCSC)[build] is the specific genome build of interest
(hg19, build37.2, GRCh37)
in a structure analogous to the igenomes package. In addition, commonly used
genomes are also available at
/fdb/gmap_post20210308/common_genomes. This path is also the
default path gmap and gsnap use to look for genome files, so a particular genome
is available there it is sufficient to provide the genome name with -d
genome_name to any of the tools without also specifying the genome
directory with -D.
Performance considerations
Gsnap throughput was measured using different numbers of threads with an
ENCODE data set (100nt single ended RNA-Seq, mouse Purkinje cells; accession
ENCFF220BOE; biosample
ENCSR758TAD). Discovery of novel splice sites was disabled but known
splice sites were provided as input. The input file was compressed and stored
on local scratch space. The output was saved unmodified on local scratch space. Each
process was allocated 30GB of memory. The exact command used was
gsnap -d mm10 --nthreads=${SLURM_CPUS_PER_TASK} \
--use-shared-memory=0 -B 5 \
--novelsplicing=0 --gunzip \
--use-splicing=/lscratch/${SLURM_JOBID}/mm10.iit \
--format=sam -o /lscratch/${SLURM_JOBID}/${n}.sam \
/lscratch/${SLURM_JOBID}/${fq}
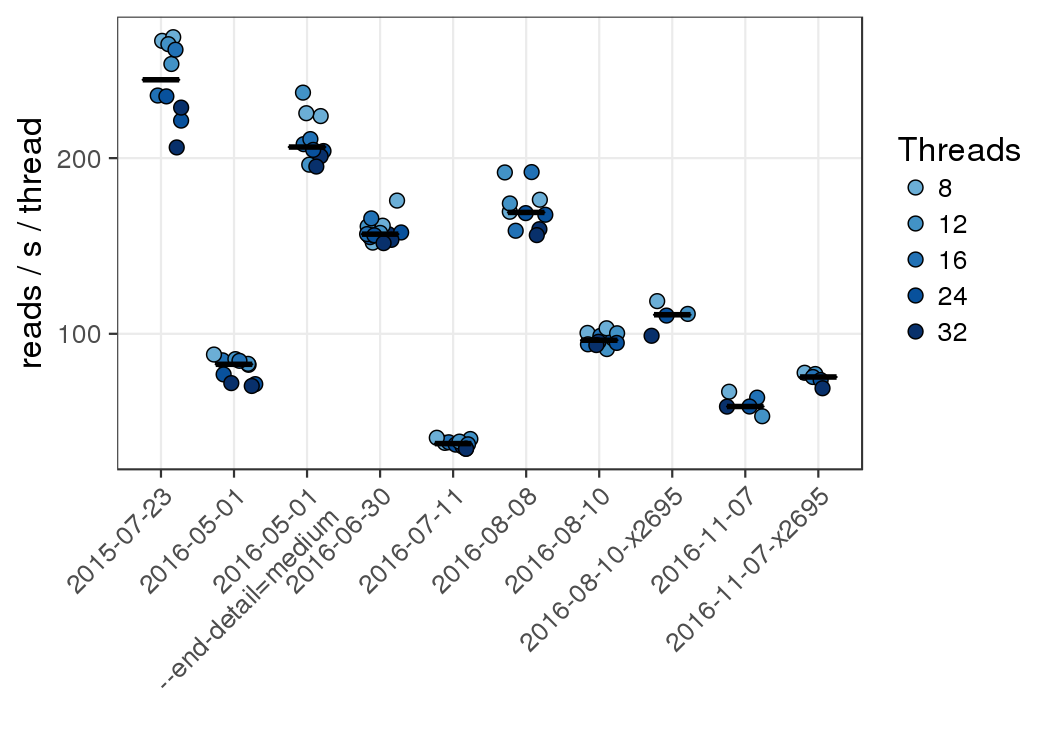
Reads aligned per second per thread for different versions of gsnap with different numbers
of threads.
The lower throughput of version 2015-05-01
with default settings is largely the result of a change in the default for one setting
( --end-detail) from medium to high which has the effect of improving
alignments in the small number of cases where the distal end of a splice or indel
contains a mismatch. Changing this version back to medium recovers most the loss
in throughput. It is therefore largely a tradeoff between throughput and sensitivity.
As of version 2016-06-30, the default reverted to medium.
Most benchmarks were run on the phase1 norm nodes (x2650; 10g). Benchmarks ending
in x2695 were run on the newer phase2 norm nodes (x2695; ibfdr).
Gsnap scales well with the number of threads all the way to 32, though the most
efficient use is probably around 24 threads with this input data set. Note that
gsnap does not start any extra processes or threads, so
--nthreads=$SLURM_CPUS_PER_TASK is appropriate unless the output is
piped into another process such as samtools. In that case the number of threads used
by the second process has to be subtracted from the number of threads given to
gsnap.
Performance was comparable when input and output were kept on the shared storage
space (/data).
On interactive session
Gmap and gsnap are multi-threaded applications. The thread count is
determined with -t / --nthreads and defaults to 1.
Map and align a single cDNA to the mouse genome. Note that the mouse genome
is in common genomes, so no path has to be provided.
[user@biowulf]$ sinteractive
[user@cn3144]$ module load gmap-gsnap
[user@cn3144]$ cd /data/$USER/test_data
[user@cn3144]$ gmap -d mm10 -A /usr/local/apps/gmap-gsnap/TEST_DATA/myc.fa
Map and align single end RNA-Seq data from encode to the mouse genome with
database of known splice junctions. Supress search for novel juctions:
[user@cn3144]$ module load samtools/1.2
[user@cn3144]$ gtf_splicesites < /fdb/igenomes/Mus_musculus/UCSC/mm10/Annotation/Genes/genes.gtf \
| iit_store -o mm10.iit
[user@cn3144]$ gsnap -d mm10 --nthreads=2 --novelsplicing=0 --use-splicing=mm10.iit --gunzip \
--format=sam \
/usr/local/apps/gmap-gsnap/TEST_DATA/ENCFF220BOE.fastq.gz \
2> test.log \
| samtools view -S -b -F4 -q 20 - \
> bam/gsnap.bam
A number of other small tools are available as part of the package.
Batch job on Biowulf
Batch scripts should use the $SLURM_CPUS_PER_TASK environment
variable to determine the number of threads to use. This variable is set by
slurm according to the number of requested cores. An example script for gsnap
might look like the following:
#! /bin/bash
# gsnap_batch.sh
set -o pipefail
set -e
function fail {
echo "$@" >&2
exit 1
}
module load samtools/1.2 || fail "could not load samtools module"
module load gmap-gsnap || fail "could not load gmap-gsnap module"
cd /data/$USER/test_data || fail "no such directory"
gtf_splicesites < /fdb/igenomes/Mus_musculus/UCSC/mm10/Annotation/Genes/genes.gtf \
| iit_store -o mm10.iit
gsnap -d mm10 --nthreads=$(( SLURM_CPUS_PER_TASK - 2 ))\
--novelsplicing=0 --use-splicing=mm10.iit --gunzip \
--format=sam \
/usr/local/apps/gmap-gsnap/TEST_DATA/ENCFF220BOE.fastq.gz \
2> test.log \
| samtools view -S -b -F4 -q 20 -@ 2 - \
> bam/gsnap.bam
The script is submitted to the batch system requesting, for example, 8 cores
and 25GB of memory:
biowulf$ sbatch -c 20 --mem=30g gsnap_batch.sh
Swarm of jobs on Biowulf
Set up a command file for two jobs, each aligning single ended reads. Note
that swarm allows line continuations.
# gsnap.swarm
cd /data/$USER/test_data \
&& gsnap -d mm10 --nthreads=$(( SLURM_CPUS_PER_TASK - 1 ))\
--novelsplicing=0 --use-splicing=mm10.iit --gunzip \
--format=sam file1.fastq.gz \
2> test1.log \
| samtools view -S -b -F4 -q 20 - \
> bam/gsnap.bam
cd /data/$USER/test_data \
&& gsnap -d mm10 --nthreads=$(( SLURM_CPUS_PER_TASK - 1 ))\
--novelsplicing=0 --use-splicing=mm10.iit --gunzip \
--format=sam file2.fastq.gz \
2> test2.log \
| samtools view -S -b -F4 -q 20 - \
> bam/gsnap2.bam
Create the known splice site database as shown above and start the jobs with swarm
biowulf$ swarm -f swarm_command_file -t 20 -g 30 \
--module samtools/1.2 gmap-gsnap
Interactive job on Biowulf
Interactive work requiring significant resources should be carried out on
interactive compute nodes, not on the head node or [user@cn3144]. Interactive nodes are
allocated with sinteractive. For example, to request a 8 core
interactive job with 25GB memory:
biowulf$ sinteractive -c 8 --mem=30g
On the interactive node, gmap and gsnap are then used essentially as decribed
above.
