From the lefse documentation:
LEfSe (Linear discriminant analysis Effect Size) determines the features (organisms, clades, operational taxonomic units, genes, or functions) most likely to explain differences between classes by coupling standard tests for statistical significance with additional tests encoding biological consistency and effect relevance.
$LEFSE_TEST_DATAAllocate an interactive session and run the program. The input format for this tool contains two rows of metadata, one row of sample ids, and a microbial abundance table.
[user@biowulf]$ sinteractive --gres=lscratch:10 --cpus-per-task=2
salloc.exe: Pending job allocation 46116226
salloc.exe: job 46116226 queued and waiting for resources
salloc.exe: job 46116226 has been allocated resources
salloc.exe: Granted job allocation 46116226
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3144 are ready for job
[user@cn3144]$ cd /lscratch/$SLURM_JOB_ID
[user@cn3144]$ module load lefse
[user@cn3144]$ cp ${LEFSE_TEST_DATA:-none}/hmp_aerobiosis_small.txt .
[user@cn3144]$ head hmp_aerobiosis_small.txt | cut -f1-4
oxygen_availability High_O2 Mid_O2 Low_O2
body_site ear oral gut
subject_id 158721788 158721788 159146620
Archaea|Euryarchaeota|Methanobacteria|Methanobacteriales|Methanobacteriaceae|Methanobrevibacter 2.96541e-06 5.08937e-06 4.93921e-06
Bacteria 0.999994 0.99999 0.99999
Bacteria|Acidobacteria 5.0412e-05 8.65194e-05 8.39666e-05
Bacteria|Acidobacteria|Acidobacteria_Gp10|Gp10 2.96541e-06 5.08937e-06 4.93921e-06
Bacteria|Acidobacteria|Acidobacteria_Gp11|Gp11 2.96541e-06 5.08937e-06 4.93921e-06
Bacteria|Acidobacteria|Acidobacteria_Gp16|Gp16 2.96541e-06 5.08937e-06 4.93921e-06
Bacteria|Acidobacteria|Acidobacteria_Gp17|Gp17 2.96541e-06 5.08937e-06 4.93921e-06
[user@cn3144]$ lefse_format_input.py hmp_aerobiosis_small.txt hmp_aerobiosis_small.in\
-c 1 -s 2 -u 3 -o 1000000
[user@cn3144]$ lefse_run.py hmp_aerobiosis_small.in hmp_aerobiosis_small.res
f significantly discriminative features: 51 ( 131 ) before internal wilcoxon
Number of discriminative features with abs LDA score > 2.0 : 51
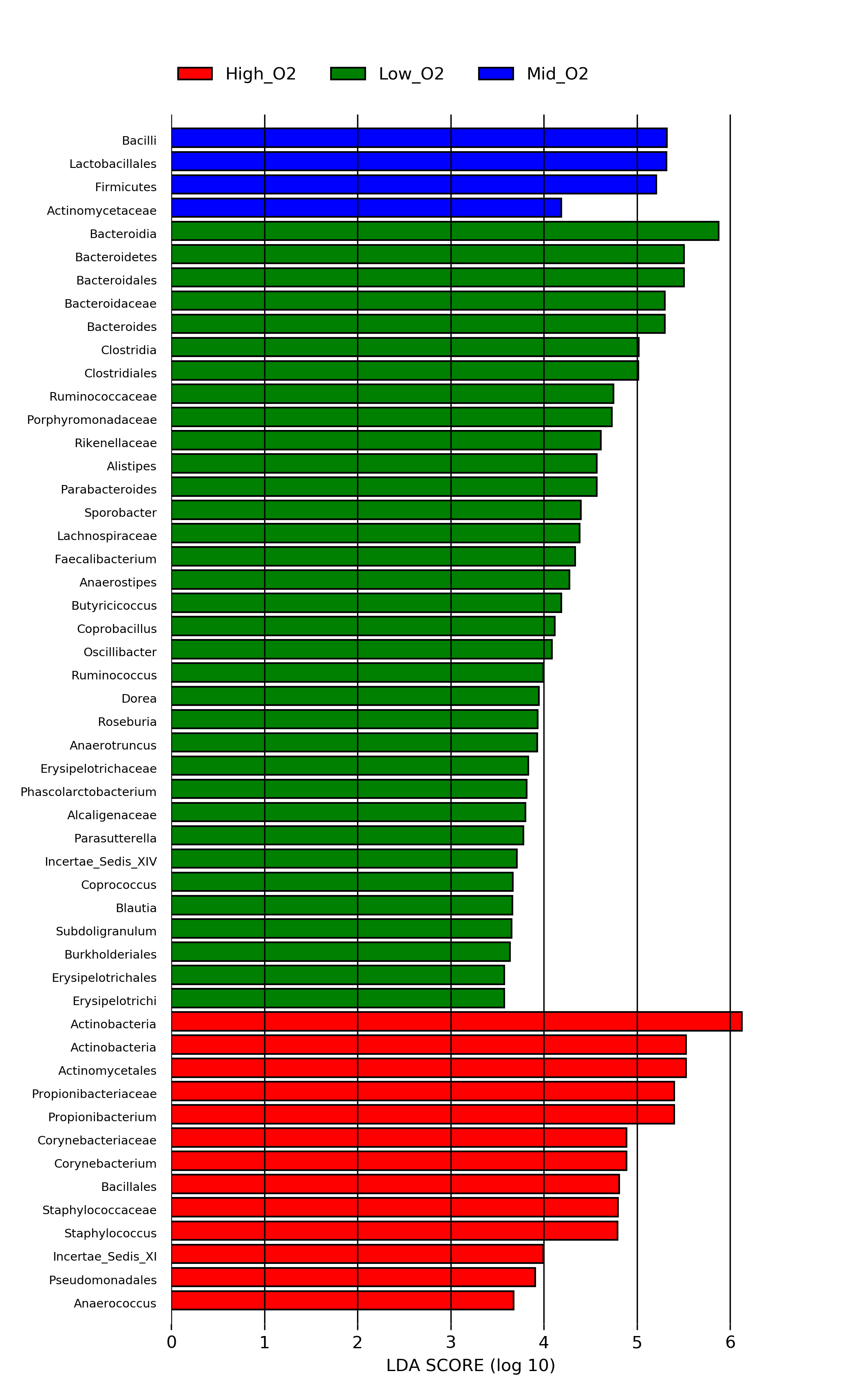
Then plot the LDA scores with
[user@cn3144]$ plot_res.py hmp_aerobiosis_small.res hmp_aerobiosis_small.png --format png --dpi=300

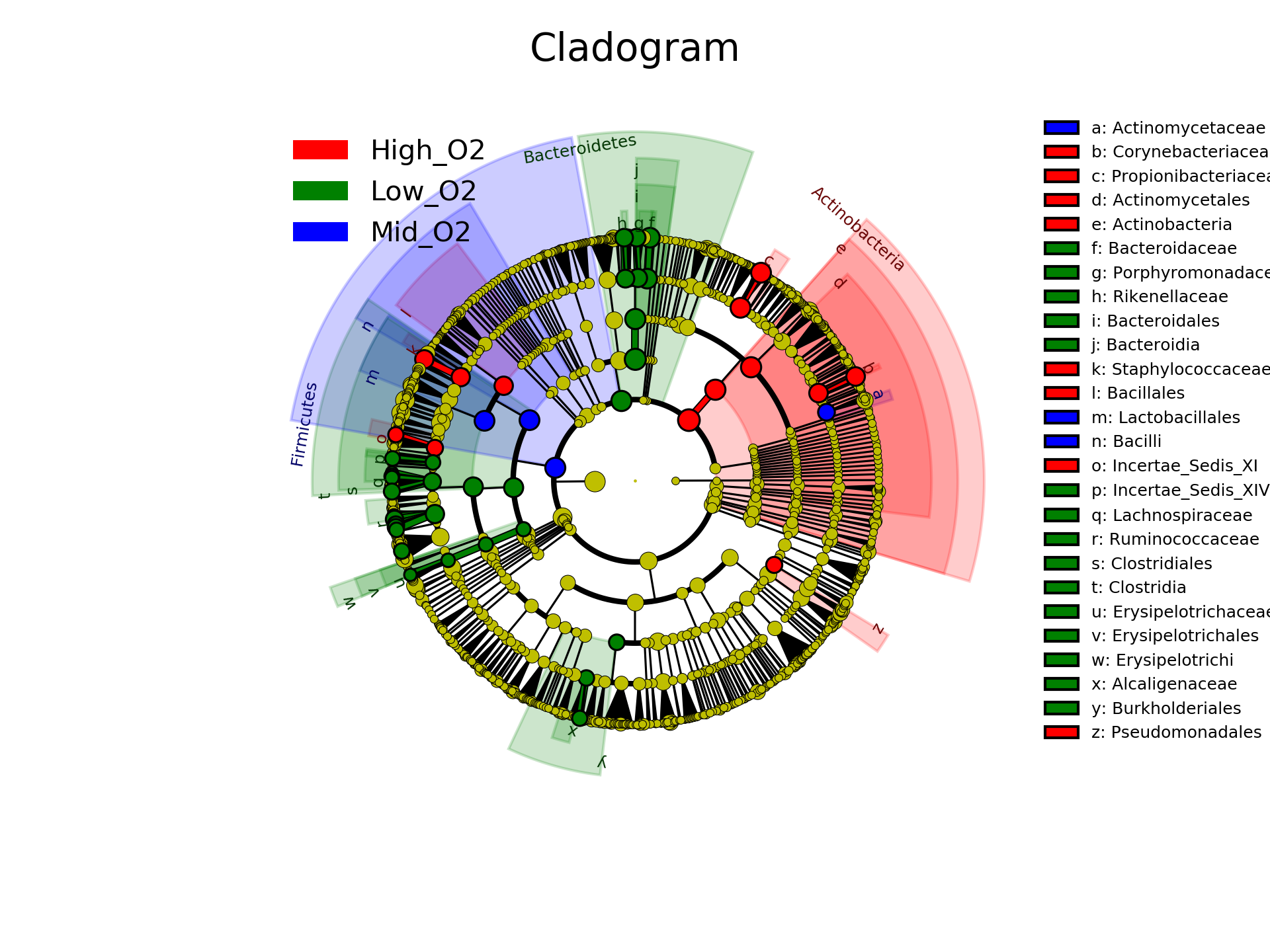
Or as a cladogram:
[user@cn3144]$ plot_cladogram.py hmp_aerobiosis_small.res hmp_aerobiosis_small.cladogram.png --format png --dpi 300
Copy results back from lscratch and exit
[user@cn3144]$ mkdir -p /data/$USER/lefse_results [user@cn3144]$ mv ./* /data/$USER/lefse_results [user@cn3144]$ exit salloc.exe: Relinquishing job allocation 46116226 [user@biowulf]$
Create a batch input file (e.g. lefse.sh), which uses the input file 'lefse.in'. For example:
#!/bin/bash
module load lefse/1.1.2
lefse_format_input.py ${LEFSE_TEST_DATA:-none}/hmp_aerobiosis_small.txt hmp_aerobiosis_small.in \
-c 1 -s 2 -u 3 -o 1000000
plot_res.py hmp_aerobiosis_small.res hmp_aerobiosis_small.png --format png --dpi=300
plot_cladogram.py hmp_aerobiosis_small.res hmp_aerobiosis_small.cladogram.png --format png --dpi 300
Submit this job using the Slurm sbatch command.
sbatch --cpus-per-task=2 --mem=5g lefse.sh