SimNIBS is used to simulate non-invasive brain stimulation. Calculations of the electric field induced by transcranial magnetic stimulation (TMS) and transcranial direct current stimulation (tDCS) are supported.
$SIMNIBS_TEST_DATAmri2mesh and headreco have been replaced by charm
(see below for an example)HPC On-demand is a web-based interface allowing use of graphical applications, useful for interactive access to this application.
Copy the test data and set up your environment
[user@biowulf]$ sinteractive
salloc.exe: Pending job allocation 46116226
salloc.exe: job 46116226 queued and waiting for resources
salloc.exe: job 46116226 has been allocated resources
salloc.exe: Granted job allocation 46116226
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3144 are ready for job
[user@cn3144]$ module load simnibs
[+] Loading simnibs 4.1.0 on cn3144
[user@cn3144]$ cd /data/$USER
[user@cn3144]$ mkdir SIMNIBS_TEST
[user@cn3144]$ cd SIMNIBS_TEST
[user@cn3144]$ cp ${SIMNIBS_TEST_DATA}/* .
[user@cn3144]$ unzip simnibs4_examples.zip
The first step would usually be the construction of a 3D mesh from MRI images. Since this step is time consuming, it should be run as a batch job. You would create a batch script similar to the following, and name it, say, make-mesh.sh:
#!/bin/bash #SBATCH --job-name=charm-job #SBATCH --cpus-per-task=64 #SBATCH --mem=20g #SBATCH --time=3:00:00 cd /data/$USER/SIMNIBS_TEST module load simnibs charm --forcerun ernie org/ernie_T1.nii.gz org/ernie_T2.nii.gz
and submit it as a batch job with
[user@biowulf]$ sbatch make-mesh.sh
Once the reconstruction jobs ends, you can visually check the segmentation pointing a browser to m2m_ernie/charm_report.html. You should be able to see a report like the following:
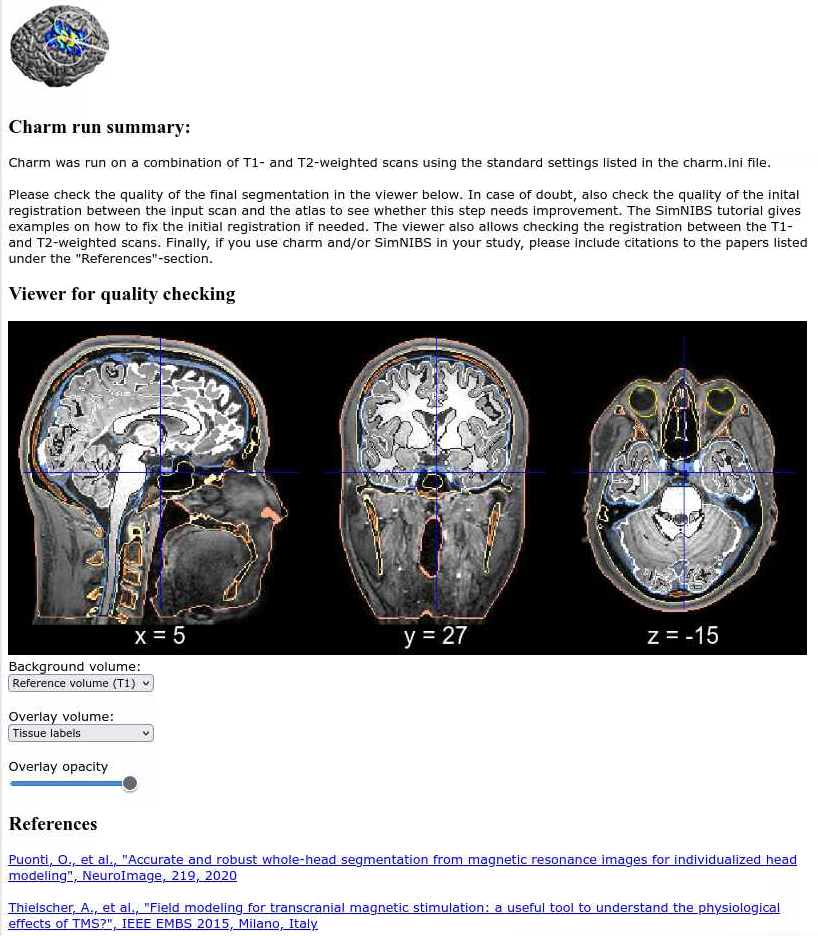
The next part of the tutorial uses the SimNIBS GUI and we will run it on an interactive node. Since this is a GUI program, using the HPC OnDemand web interface to connect to biowulf is likely to give better responsiveness.
[user@biowulf]$ sinteractive --mem=10g salloc.exe: Pending job allocation 46116226 salloc.exe: job 46116226 queued and waiting for resources salloc.exe: job 46116226 has been allocated resources salloc.exe: Granted job allocation 46116226 salloc.exe: Waiting for resource configuration salloc.exe: Nodes cn3144 are ready for job [user@cn3144]$ module load simnibs [+] Loading simnibs 4.1.0 on cn3144 [user@biowulf]$ cd /data/$USER/SIMNIBS_TEST [user@biowulf]$ simnibs_gui &
This should start a GUI-based viewer. You will find a step-by-step guide on how to set up and run simulations via the GUI at the Simnibs GUI Tutorial. However, it is useful to run simulations as batch jobs, as shown in the section below.
Create the following python script (based on the simnibs Tutorial's section on scripting simulations), and name it tms-sim.py:
import os, sys
from simnibs import sim_struct, run_simnibs
# Init session
s = sim_struct.SESSION()
# Name of mesh
s.subpath = sys.argv[1]
# Output
s.pathfem = sys.argv[2]
# Init TMS simulations
tmslist = s.add_tmslist()
# Select coil
tmslist.fnamecoil = os.path.join('legacy_and_other','Magstim_70mm_Fig8.ccd')
# Initialize coil position
pos = tmslist.add_position()
# Select coil centre
pos.centre = 'C1'
# Select coil direction
pos.pos_ydir = 'CP1'
# Don't open in gmsh
s.open_in_gmsh = False
# Launch simulation
run_simnibs(s)
Now, create a batch input file (e.g. tms-sim.sh) that will execute the python script above, using as input the subject m2m_ernie and writing the output to the directory tms-sim-out:
#!/bin/bash #SBATCH --job-name=tms-sim #SBATCH --mem=8g #SBATCH --time=2:00:00 module load simnibs/4.1.0 cd /data/$USER/SIMNIBS_TEST python tms-sim.py m2m_ernie tms-sim-out
Submit this job using the Slurm sbatch command.
sbatch tms-sim.sh
Create a swarmfile (e.g. tms-sim.swarm) that uses the script tms-sim.py above to run a simulation for 5 different subjects and writes the output to 5 different output directories:
cd /data/$USER/SIMNIBS_TEST; python tms-sim.py sub01 out1 cd /data/$USER/SIMNIBS_TEST; python tms-sim.py sub02 out2 cd /data/$USER/SIMNIBS_TEST; python tms-sim.py sub03 out3 cd /data/$USER/SIMNIBS_TEST; python tms-sim.py sub04 out4 cd /data/$USER/SIMNIBS_TEST; python tms-sim.py sub05 out5
Submit this job using the swarm command.
swarm -f tms-sim.swarm -g 8 -t 2 --module simnibswhere
| -gres=lscratch:# | Number of Gigabytes of local disk space allocated per process (1 line in the swarm command file) |
| -g # | Number of Gigabytes of memory required for each process (1 line in the swarm command file) |
| -t # | Number of threads/CPUs required for each process (1 line in the swarm command file). We
set this to auto to allocate all CPUs in each node.
|
| --module simnibs | Loads the simnibs module for each subjob in the swarm |