The weblogo package includes a tool ( seqlogo) to generate
sequence logos from aligned sequences in fastq format.
Note that weblogo changed significantly from version 2.X to 3.X. The commands listed here are for versions 3.X
Allocate an interactive session and run the program.
Sample session (user input in bold):
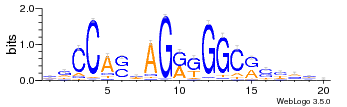
[user@biowulf]$ sinteractive salloc.exe: Pending job allocation 46116226 salloc.exe: job 46116226 queued and waiting for resources salloc.exe: job 46116226 has been allocated resources salloc.exe: Granted job allocation 46116226 salloc.exe: Waiting for resource configuration salloc.exe: Nodes cn3144 are ready for job [user@cn3144 ~]$ module load weblogo [+] Loading weblogo 3.6 [user@cn3144 ~]$ cp $WEBLOGO_DATA/* . [user@cn3144 ~]$ weblogo -f ctcf.fa -F png -o ctcf.png [user@cn3144 ~]$ exit salloc.exe: Relinquishing job allocation 46116226 [user@biowulf ~]$Visualization of the output image ctcf.png:

Create a batch script similar to the following example:
#! /bin/bash #SBATCH --mem=100m #SBATCH --time=5 # this file is seqlogo_job.sh module load weblogo || exit 1 cp $WEBLOGO_DATA/* . weblogo -f ctcf.fa -F png -o ctcf.png
Submit to the queue with sbatch:
biowulf$ sbatch seqlogo_job.sh
Create a swarm command file similar to the following example:
# this file is seqlogo_jobs.swarm seqlogo -f site1.fa -F eps -o site1 -abcMnY seqlogo -f site2.fa -F eps -o site2 -abcMnY seqlogo -f site3.fa -F eps -o site3 -abcMnY seqlogo -f site4.fa -F eps -o site4 -abcMnY
And submit to the queue with swarm
biowulf$ swarm -f seqlogo_jobs.swarm --time=5