I-TASSER on Biowulf
I-TASSER (Iterative Threading ASSEmbly Refinement) is a hierarchical approach to protein structure and function prediction.
- Structural templates are first identified from the
PDB by multiple threading approach
- LOMETS; full-length atomic models are then constructed by iterative template fragment assembly simulations.
- Finally, function inslights of the target are derived by threading the 3D models through protein function database BioLiP.
I-TASSER (as 'Zhang-Server') was ranked as the No 1 server for protein structure prediction in recent CASP7, CASP8, CASP9, CASP10, and CASP11 experiments.
It was also ranked as the best for function prediction in CASP9. The server is in active development with the goal to provide the most accurate structural and function predictions using state-of-the-art algorithms.
I-TASSER was developed in the Zhang lab at the University of Michigan. I-TASSER website
On the NIH HPC systems, the runI-TASSER.pl script has been modified to internally contain the locations of the I-TASSER libraries. Thus, the parameter --libdir need not be provided
in your command line.
The I-TASSER libraries are updated on our systems every Friday morning.
Single-sequence job on Biowulf
The following examples use the sample data that is provided with I-TASSER. This can be copied from $ITASSER_HOME/example, as is done in the following batch script.
Important:
- The input query sequence MUST be called seq.fasta.
- Each I-TASSER run will create files with the same filenames, so it is very important that each I-TASSER run be performed in a unique directory.
- The I-TASSER 'database' consists of a massive number of small files. Likewise, the temporary files during the program run consist of many small files. Reading and writing these files puts a heavy load on the filesystems. Therefore it is best to copy the database to local scratch on the node at the beginning of the job, and read/write to local scratch thereafter. In our tests, this process took 30% less time than reading/writing from the central filesystems. For the test job:
Reading/writing from central filesystems: 33 hrs runtime
Reading/writing from local scratch on node: 23 hrs runtime
The module will copy the I-TASSER database to local scratch, and also use local scratch as the temporary directory. However, it is important to _request_ local scratch as part of your sbatch command, as in the example below.
Create a batch input file (e.g. itasser.sh). For example:
#!/bin/sh
set -e
module load I-TASSER
datadir="/data/$USER/I-TASSER/example"
export TMPDIR="/lscratch/$SLURM_JOB_ID"
runI-TASSER.pl \
-runstyle gnuparallel \
-LBS true \
-EC true \
-GO true \
-seqname example \
-datadir "$datadir" \
-light true \
-outdir "$datadir" \
&& file2html.py "$datadir"
Submit this job using the Slurm sbatch command.
sbatch --time=1-00:00:00 --cpus-per-task=14 --mem=170g --gres=lscratch:100 itasser.sh
where
| --time=1-00:00:00 | Set a walltime max of 1 day
|
| --cpus-per-task=14 | Allocate 14 CPUs. Note: Blast really doesn't scale beyond 4 CPUs, but I-TASSER will run 14 simulations at a point in the analysis.
so there's no point increasing this value.
|
| --mem=170g | Allocate 170 GB of memory. The memory requirement was determined via a test job.
|
| --gres=lscratch:100 | Allocate 100 GB of local disk
|
Swarm of jobs on Biowulf
To run I-TASSER with a large number of input sequences, it is most convenient to use the Biowulf swarm utility. Note that for each input sequence,
I-TASSER will write output files with the same names, so that each run MUST be performed in a unique directory.
Start by setting up the directories for each run, with a single sequence file called seq.fasta in each directory. Your initial directory tree might look like this:
---- /data/$USER/ ---------
|-- I-TASSER
| |-- seq0
| | |-- seq.fasta
| |-- seq1
| | |-- seq.fasta
| |-- seq2
| | |-- seq.fasta
| |-- seq3
| | |-- seq.fasta
| |-- seq4
| |-- seq.fasta
The outputs for each sequence should go into the directories seq0, seq1....
Create a swarm command file in your top-level I-TASSER directory (e.g. /data/$USER/I-TASSER) along the following lines:
# this file is called itasser.swarm
export TMPDIR="/lscratch/$SLURM_JOB_ID"; runI-TASSER.pl -runstyle gnuparallel -LBS true -EC true -GO true -seqname seq0 -datadir ./seq0 -light true -outdir ./seq0 && file2html.py ./seq0
export TMPDIR="/lscratch/$SLURM_JOB_ID"; runI-TASSER.pl -runstyle gnuparallel -LBS true -EC true -GO true -seqname seq1 -datadir ./seq1 -light true -outdir ./seq1 && file2html.py ./seq1
export TMPDIR="/lscratch/$SLURM_JOB_ID"; runI-TASSER.pl -runstyle gnuparallel -LBS true -EC true -GO true -seqname seq2 -datadir ./seq2 -light true -outdir ./seq2 && file2html.py ./seq2
export TMPDIR="/lscratch/$SLURM_JOB_ID"; runI-TASSER.pl -runstyle gnuparallel -LBS true -EC true -GO true -seqname seq3 -datadir ./seq3 -light true -outdir ./seq3 && file2html.py ./seq3
export TMPDIR="/lscratch/$SLURM_JOB_ID"; runI-TASSER.pl -runstyle gnuparallel -LBS true -EC true -GO true -seqname seq4 -datadir ./seq4 -light true -outdir ./seq4 && file2html.py ./seq4
Submit this swarm with:
swarm -t 8 -g 170 -f itasser.swarm --time=2-00:00:00 --gres=lscratch:100 --module I-TASSER/5.1
This swarm command will allocate 8 CPUs (on which the blastpgp runs will be multi-threaded) and 170 GB of memory to each I-TASSER run. The walltime for each run has been set to 2 days. 100 GB of local disk
will be allocated for each run, and the runI-TASSER.pl script has been modified to copy the I-TASSER data and tmp files to local disk.
Typical jobload for a swarm of runs (running in serial mode, v4.4):
[user@biowulf ~]$ jobload -u user
JOBID TIME NODES CPUS THREADS LOAD MEMORY
Elapsed / Wall Alloc Active Used/Alloc
16062491_4 00:19:32 / 2-00:00:00 cn0228 4 4 100% 22.3 GB/60.0 GB
16062491_0 00:20:17 / 2-00:00:00 cn0059 4 1 25% 542.1 MB/60.0 GB
16062491_1 00:20:16 / 2-00:00:00 cn0293 4 4 100% 13.8 GB/60.0 GB
16062491_2 00:20:16 / 2-00:00:00 cn0243 4 4 100% 35.0 GB/60.0 GB
16062491_3 00:20:16 / 2-00:00:00 cn0074 4 1 25% 30.0 GB/60.0 GB
USER SUMMARY
Jobs: 5 Nodes: 5 CPUs: 20 Load Avg: 70%%
Interactive job on Biowulf
Allocate an interactive session and run I-TASSER on there. Note that the max walltime for an interactive job is 36 hrs, so while this interactive job is succesful for
the test job below, it might not be sufficient for longer sequences where the I-TASSER runs will be longer.
Sample session:
[user@biowulf ~]$ sinteractive --cpus-per-task=14 --mem=170g --gres=lscratch:100
salloc.exe: Pending job allocation 39611486
salloc.exe: job 39611486 queued and waiting for resources
salloc.exe: job 39611486 has been allocated resources
salloc.exe: Granted job allocation 39611486
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn3535 are ready for job
[user@cn3535 ~]$ export TMPDIR="/lscratch/$SLURM_JOB_ID"
[user@cn3535 ~]$ cd /data/user/I-TASSER
[user@cn3535 I-TASSER]$ module load I-TASSER
[user@cn3535 I-TASSER]$ cp -r $ITASSER_HOME/example/seq.fasta .
[user@cn3535 I-TASSER]$ runI-TASSER.pl -runstyle gnuparallel -LBS true -EC true -GO true -seqname seq.fasta -datadir . -light true -outdir ./example
Your setting for running I-TASSER is:
-pkgdir = /usr/local/apps/I-TASSER/5.1
-libdir = /lscratch/39611486/ITLIB.ZWE5/I-TASSER
-java_home = /usr
-seqname = seq.fasta
-datadir = /spin1/users/user/I-TASSER/example
-outdir = /spin1/users/user/I-TASSER/example
-runstyle = gnuparallel
-homoflag = real
-idcut = 1
-ntemp = 20
-nmodel = 5
-light = true
-hours = 6
-LBS = true
-EC = true
-GO = true
Username:
Tmpdir:
1. make seq.txt and rmsinp
Your protein contains 143 residues:
> seq.fasta
MYQLEKEPIVGAETFYVDGAANRETKLGKAGYVTNRGRQKVVTLTDTTNQKTELQAIYLA
LQDSGLEVNIVTDSQYALGIITQWIHNWKKRGWPVKNVDLVNQIIEQLIKKEKVYLAWVP
AHKGIGGNEQVDKLVSAGIRKVL
2.1 run Psi-blast
2.2 Predict secondary structure with PSSpred...
2.3 Predict solvent accessibility...
2.4 run pairmod
2.4.1 Use all templates
2.4.2 running pair ................
FORTRAN STOP
30000 6379375 total lib str & residues
number of observations 27.03459 1658491.
pair done
3.1 do threading
start gnuparallel threading PPAS
start gnuparallel threading dPPAS
start gnuparallel threading dPPAS2
start gnuparallel threading Env-PPAS
start gnuparallel threading MUSTER
start gnuparallel threading wPPAS
start gnuparallel threading wdPPAS
start gnuparallel threading wMUSTER
FORTRAN STOP
3.2 make restraints
[....etc...]
[user@cn3535 I-TASSER]$ file2html.py ./example
PyMOL(TM) Molecular Graphics System, Version 1.8.0.0.
Copyright (c) Schrodinger, LLC.
All Rights Reserved.
Created by Warren L. DeLano, Ph.D.
PyMOL is user-supported open-source software. Although some versions
are freely available, PyMOL is not in the public domain.
[...]
Viewing the HTML Report
You can review your results directly from your desktop web browser if you locally mount your HPC storage via hpcdrive.
There is no need to open a shell session or copy files to do so.
Benchmarks
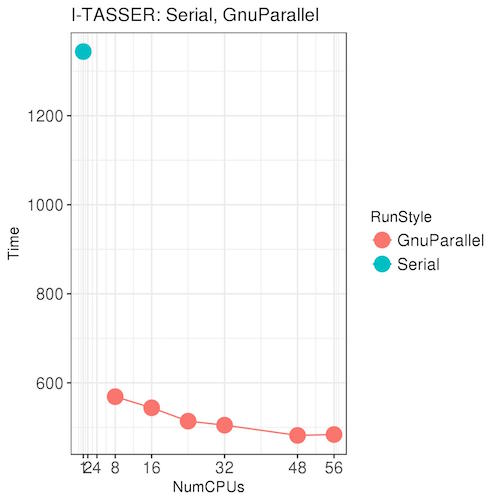
| The test job in the graph on the left shows the advantage of running I-TASSER using the option -runstyle gnuparallel. With 8 CPUs and Gnu Parallel,
the job runs almost 3x faster than the serial run on a single CPU.
Using more than 8 CPUs with Gnu Parallel can provide an additional but much smaller boost, but remember that requesting more CPUs is likely
to cause your job to take longer to get scheduled by the batch system. However, if your job requires most of the memory on a node, you might as well request all the CPUs on that node as well.
Recommendation: if your I-TASSER jobs require 150 GB of memory or less, submit to 8 CPUs. If your I-TASSER jobs require close to 250 GB of memory, request all 56 CPUs.
|
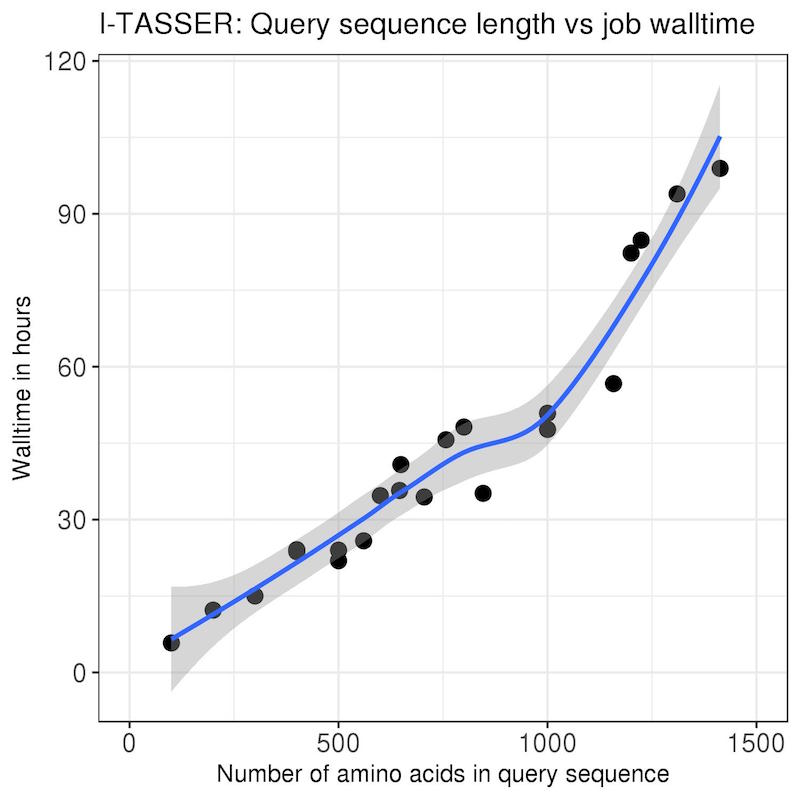
| The benchmarks on the left were standard I-TASSER runs with --runstyle gnuparallel and query sequences of varying lengths
pulled from the PDB. All jobs were run on x2680 nodes on the norm partition with 8 CPUs and 170 GB of memory.
This should give you some idea of how much walltime to request when you submit your jobs. Make sure you add a time buffer of at least 6 hrs, and monitor your job to check that it's not running out of time. You can set
#SBATCH --mail-type=TIME_LIMIT_50,TIME_LIMIT_80,END
in your batch script to send you email alerts when your job reaches 50% and 80% of the walltime limit. If you discover your job is running out of time, use newwall to add more walltime.
|
Documentation

