MATLAB on NIH HPC Systems
Quick Links
On this page
Related
|
Description

MATLAB integrates mathematical computing and data visualization in a powerful language to provide a flexible environment for technical computing. The open architecture makes it easy to use MATLAB and its companion products to explore data and create custom tools. MATLAB users benefit from a growing community, pre-written toolboxes containing popular algorithms and analyses, and an integrated development environment (IDE) for writing and debugging code.
Web sites
| |
This application can be utilized interactively via the
HPC OnDemand web interface.
Important Notes
back to top
Licenses are checked out automatically when you use a MATLAB or its toolboxes. Licenses are returned when your sinterative session ends. There is no maximum number of MATLAB sessions a single user can run. However, as more than one instance of Matlab can run within a single interactive session, users must make sure they request increased resource allocation for their interactive session if more than one Matlab instance will be launched.
As with other applications on the HPC systems, MATLAB is managed using environment modules. To see which versions of MATLAB and third-party toolboxes are available, type:
[user@cn1234 ~]$ module avail matlab
----------------------------------------------------------------
matlab-eeglab/2022.0 matlab-spm/12.7219 matlab/2022a matlab/2023a
Module defaults are chosen based on Find First Rules due to Name/Version/Version modules found in the module tree.
See https://lmod.readthedocs.io/en/latest/060_locating.html for details.
If the avail list is too long consider trying:
"module --default avail" or "ml -d av" to just list the default modules.
"module overview" or "ml ov" to display the number of modules for each name.
Use "module spider" to find all possible modules and extensions.
Use "module keyword key1 key2 ..." to search for all possible modules matching any of the "keys".
[user@cn1234 ~]$
Please note that, while we have installed several third-party toolboxes in the past, the best place to install third-party toolboxes is your data directory. Please email us at staff@hpc.nih.gov if you require assistance with third-party toolbox installation.
To select a module, type:
[user@cn1234 ~]$ module load matlab/[ver]
where [ver] is an optional version specification. Without [ver] the default MATLAB version is loaded.
Please note that, for 2017a and later, the complete set of third-party toolboxes will not be automatically loaded unless you choose the matlab.all module or load them individually.
To load a third-party toolbox, like SPM, for use with MATLAB >= 2017a, do:
[user@cn1234 ~]$ module load matlab-spm
If you have already begun a MATLAB session, there is no need to restart.
The loading can be accomplished using the nih_matmod function:
>> nih_matmod avail
eeglab/2022.0 spm/12.7219
Module defaults are chosen based on Find First Rules due to Name/Version/Version modules found in the module tree.
See https://lmod.readthedocs.io/en/latest/060_locating.html for details.
If the avail list is too long consider trying:
"module --default avail" or "ml -d av" to just list the default modules.
"module overview" or "ml ov" to display the number of modules for each name.
Use "module spider" to find all possible modules and extensions.
Use "module keyword key1 key2 ..." to search for all possible modules matching
any of the "keys".
>> nih_matmod load spm
Use help nih_matmod from within MATLAB for complete usage information.
Interactive sessions on Biowulf
Because MATLAB is not permitted on the Biowulf login node, users must request resources on a compute node.
First, establish an ssh or X-Windows connection to the Biowulf login node. When running an X-Windows session on Biowulf, remember to use the -X or -Y option with your ssh command.
Then use the sinteractive command to request resources on a compute node:
[user@biowulf ~]$ sinteractive -c 8 --mem=10g
salloc.exe: Pending job allocation 15323416
salloc.exe: job 15323416 queued and waiting for resources
salloc.exe: job 15323416 has been allocated resources
salloc.exe: Granted job allocation 15323416
salloc.exe: Waiting for resource configuration
salloc.exe: Nodes cn1640 are ready for job
[user@cn1640 ~]$
By default, the sinteractive command only allocates a few cpus and a small amount of memory. You can request more cpus and memory with the --cpus-per-task=ncpus and mem=GBg options respectively. See sinteractive -h for a complete list of options.
Graphical Interactive sessions
To run the MATLAB integrated development environment (IDE), we recommend the HPC OnDemand web interface.
HINT: Debugging Graphic Sessions
If you have trouble displaying the MATLAB IDE, ensure that your client is working properly by typing 'xclock' in the shell.
After connecting to a compute node, load the MATLAB module.
[user@cn1234 ~]$ module load matlab
[user@cn1234 ~]$ matlab&
Including & in your command above allows you to continue using the terminal while the MATLAB application is running. You should now see a MATLAB splash screen followed by the IDE:

Interactive shell sessions
It is also possible to run MATLAB interactively on the command-line, without the IDE. This would be useful if you do not wish to use X-Windows.
[user@cn1234 ~]$ module load matlab
[user@cn1234 ~]$ matlab -nodisplay
< M A T L A B (R) >
Copyright 1984-2021 The MathWorks, Inc.
R2021a Update 3 (9.10.0.1684407) 64-bit (glnxa64)
May 27, 2021
To get started, type doc.
For product information, visit www.mathworks.com.
>> quit
[user@cn1234 ~]$
Parallel Computing within a single compute node
There are a few ways in which you can parallelize your Matlab code within the same compute node. For example, in the cross product of a matrix, Matlab can do implicit parallelism by using more than one thread. The number of threads used by such operations is controlled by Matlab's maxNumCompThreads and it is set automatically to the number of CPUs allocated to your batch or interactive job. You can also set its value by executing maxNumCompThreads(new_value) within Matlab. Below is the average runtime of the cross product of a matrix with different values of maxNumCompThreads.
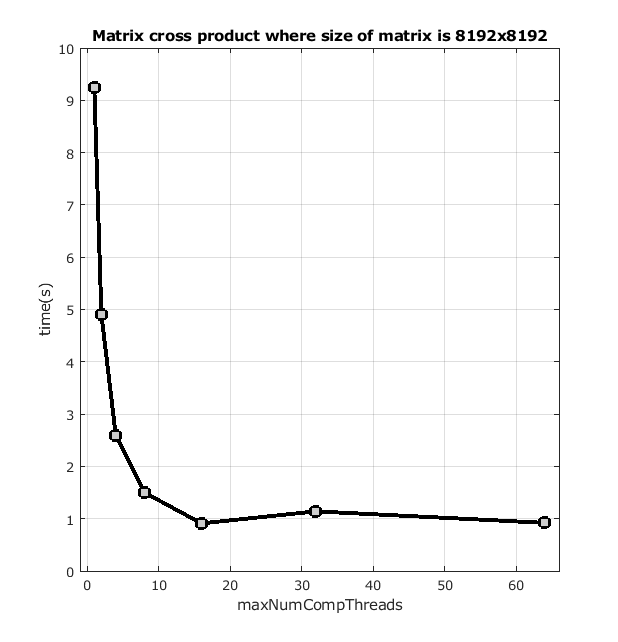
back to top
MATLAB commands and variables hardcoded into shell script
The simplest (but perhaps least useful) way to run a MATLAB job in the background would be to hardcode variables and MATLAB commands directly into a script. Even though this method is not very practical it serves as a useful example to MATLAB users unfamiliar with bash scripting:
#!/bin/bash
# this file is called hyp1.sh
module load matlab
matlab -nojvm<<-EOF
a = 3;
b = 4;
H = sqrt(a^2 + b^2)
exit
EOF
You could run this job in the background of an interactive session like so:
[user@cn0001 ~]$ ./hyp1.sh > hyp1_output 2>&1 &
HINT: Permission denied error
If you receive an error such as this:
bash: ./hyp1.sh: Permission denied
Use the chmod command to make the file executable, like so:
[user@cn0001]$ chmod 755 hyp1.sh
In this case, the output will be redirected to hyp1_output. The computation will proceed in the background allowing the you to run another session of MATLAB interactively or run other MATLAB programs in the background. If you use this strategy to run a few instances of MATLAB in parallel you must be careful not to overload your allocated CPUs.
HINT:`EOF' warning
If a message similar to the following is found in your output file, make sure to remove any trailing whitespace from both of the EOF lines.
/var/spool/slurm/slurmd/job48226/slurm_script: line 10: warning: here-document at line 5 delimited by end-of-file (wanted `EOF')
Passing MATLAB variables to a shell script
You can set up a batch script that contains MATLAB commands and pass variables to the job at the time of submission. This might be useful for very simple MATLAB analyses. For example:
#!/bin/bash
# this file is called hyp2.sh
a=$1
b=$2
echo "a is $a, b is $b";
module load matlab
matlab -nojvm<<-EOF
H = sqrt($a^2 + $b^2)
exit
EOF
In this script the values of a and b are expected to be passed from the command line. This is accomplished like so:
[user@cn0001 ~]$ ./hyp2.sh 22 5 > hyp2_output 2>&1 &
Similar to the previous example, the output is directed to hyp2_output.
MATLAB functions in shell scripts
Of course it is possible to write your own functions and call them in a shell script using the same syntax as above. Let's assume you write a function and save it in hyp.m
function H = hyp(a,b)
if ischar(a), a = str2double(a); end
if ischar(b), b = str2double(b); end
H = sqrt(a^2 + b^2)
To run the code in the background, you could write a script like so:
#!/bin/bash
# this file is called hyp3.sh
a=$1
b=$2
module load matlab
matlab -nojvm<<-EOF
cd /full/path/to/hyp.m
hyp($a,$b);
exit
EOF
Similar to the last example you would run this code in the backround with:
[user@cn0001 ~]$ ./hyp3.sh 8 4 > hyp3_output 2>&1 &
This method allows you to develop analyses of any complexity in MATLAB (rather than hardcoding commands into shell scripts) and run them in the background of an interactive session.
One common approach is to write a MATLAB function that accepts a file name as input, loads the file (containing all variables and data), performs some analysis, and then saves the analyzed data to a new .mat file. This minimizes the complexity of the input that must be supplied through a shell script. In this case, the _output file will only be used for diagnosing a debugging problems, because the output will be saved to a new .mat file specified by the user in the function.
Using the GPUs with Matlab
Several toolboxes can use GPU resources. Some of those toolboxes are: Statistics and Machine Learning, Image processing, Deep Learning, Computer Vision, Signal Processing, Wavelet, Curve Fitting, Parallel Computing. Many functions in the Deep Learning Toolbox use GPU resources automatically (see Matlab's Deep Learning with GPUs). For all other toolboxes users must explicitly elect to use GPU resources by passing a gpuArray data structure to a given function. For further details on how to use GPU with Matlab, please see Matlab's Help Center section on how to Run Matlab Functions on a GPU.
Parallel Computing Toolbox
The MATLAB Parallel Computing Toolbox enables you to develop distributed and parallel MATLAB applications and execute them using multiple cores in a single node or to utilize the graphical processing units (GPUs) of a properly equipped machine.
Please NOTE:
1. In it's current configuration, the Parallel Computing Toolbox does not scale beyond a single node. This will allow your job to run up to 16 times faster on the norm partition. This may be sufficient for many jobs. But this toolbox will not allow you to run jobs on multiple nodes. To run jobs of larger scale, use sbatch or swarm.
2. Within MATLAB and in online documentation this toolbox is referred to as the Parallel Computing Toolbox. Within the licensing software it is referred to as the Distributed Processing Toolbox. There is a related Mathworks product that is not currently installed on our systems that extends the functionality of the Parallel Computing Toolbox called the Distributed Computing Server. This has been the source of confusion.
See MATLAB Parallel Computing Toolbox for examples on using the toolbox on biowulf.
back to top
Make sure to use the -batch flag each slurm job, as shown below.
Long running jobs, or jobs that are "embarrassingly parallel" should be submitted to the batch system. To submit matlab code as a batch job, you should write another a shell script as follows:
#!/bin/bash
# this file is called hyp4.sh
#SBATCH --job-name=hypotenuse4
#SBATCH --mail-type=BEGIN,END
module load matlab
cd /data/$USER/my-matlab-scripts
matlab -batch hyp(3,4)
Note the optional inclusion of #SBATCH options that set the job name and email alerts. You would then submit the job like so:
[user@biowulf ~]$ sbatch /data/user/hyp4.sh
For more information on using the sbatch command for SLURM, visit the Job Submission section of the Biowulf User Guide.
back to top
Make sure to use the -batch flag each slurm job, as shown below.
The swarm program is a convenient way to submit large numbers of jobs. With swarm you can run several instances of your compiled MATLAB code distributed across the cluster. You create a swarm command file containing a single line for each independent job. The swarm program will then package up the commands into batch jobs and submit them to Slurm for you.
To run a swarm based on the example above, create a swarm command file named hyp.swarm with each line containing a single command:
#SWARM --job-name=hypothenuse4
cd /data/$USER/my-matlab-scripts; matlab -batch hyp(3,4)
cd /data/$USER/my-matlab-scripts; matlab -batch hyp(5,6)
cd /data/$USER/my-matlab-scripts; matlab -batch hyp(7,8)
Submit this file to the batch system with the command:
[user@biowulf ~]$ swarm -f hyp.swarm --module=matlab
Please note that, with this type of swarm, subjob packing (-p option) should not be used. Bundling (-b), however, would work as expected.



