June 2020
Benchmarks of Acemd v3 test suite on Biowulf GPU nodes. The test suite is provided with the Acemd package, and is available on Biowulf in /usr/local/apps/acemd/acemd3_examples/.
Hardware:
The runs were performed with varying numbers of CPUs. In most cases, 2 CPUs per GPU gave the best performance for a given number of GPUs, therefore all results presented below were done with 2 CPUs per GPU.
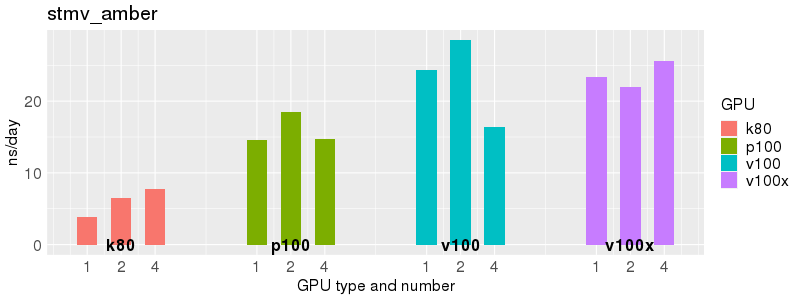
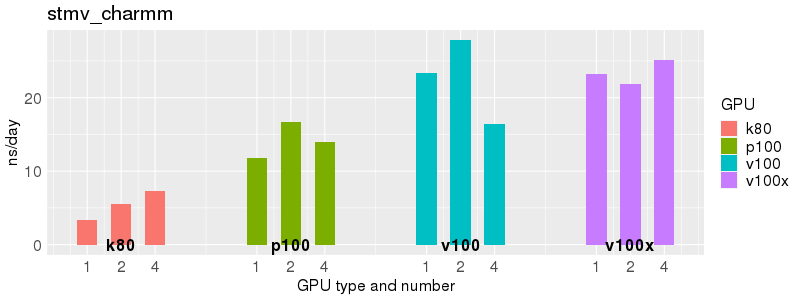
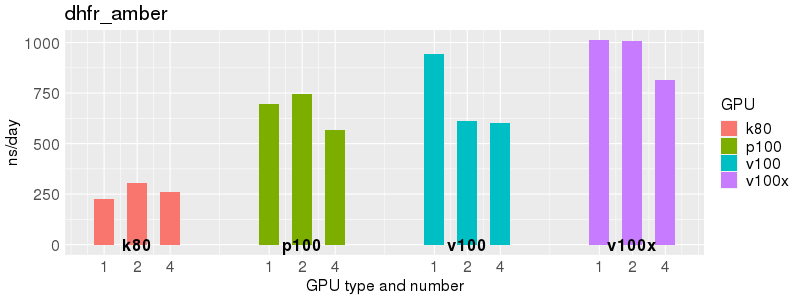
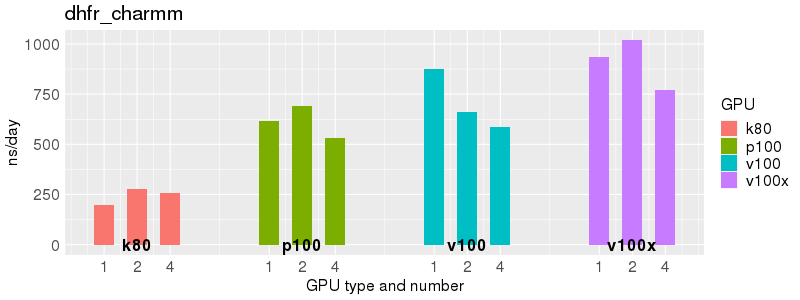
Based on these benchmarks, there is a significant performance advantage to the v100 and v100x over the p100 and k80 GPUs. However, in most cases there is no advantage to using more than 1 GPU of any type. Before allocating multiple GPUs for your own Acemd jobs, please run your own benchmarks to verify that the additional GPUs are providing a performance improvement for your jobs. (and please send any interesting results to staff@hpc.nih.gov).
Number of atoms: 23,558. Box size: 62.23 Å ✕ 62.23 Å ✕ 62.23 Å


Number of atoms: 90,906. Box size: 142.09 Å ✕ 83.34 Å ✕ 78.68 Å
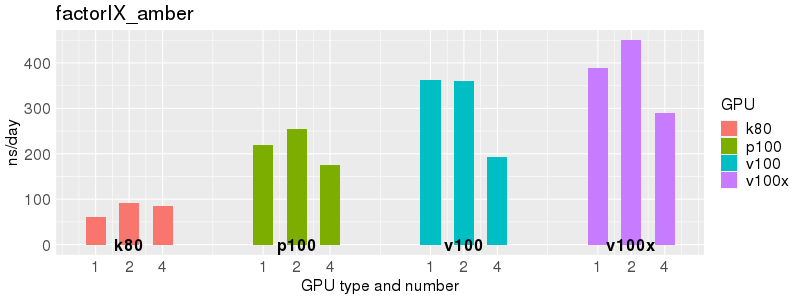
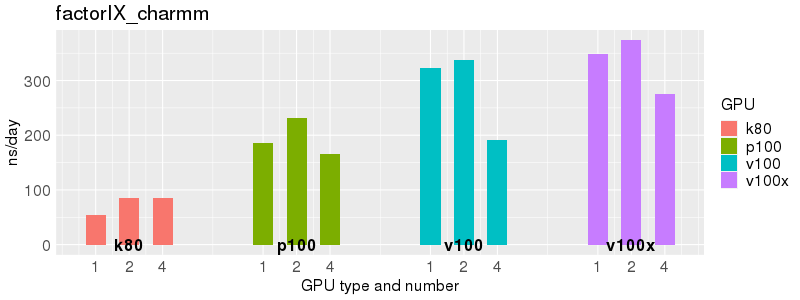
Number of atoms: 1,067,095. Box size: 221.2 Å ✕ 223.2 Å ✕ 224.5 Å